- Biology Article
Table of Contents

What are Lipids?
Properties of lipids, lipid structure, classification of lipids, types of lipids, examples of lipids, lipids definition.
“Lipids are organic compounds that contain hydrogen, carbon, and oxygen atoms, which form the framework for the structure and function of living cells.”

These organic compounds are nonpolar molecules, which are soluble only in nonpolar solvents and insoluble in water because water is a polar molecule. In the human body, these molecules can be synthesized in the liver and are found in oil, butter, whole milk, cheese, fried foods and also in some red meats.
Let us have a detailed look at the lipid structure, properties, types and classification of lipids.
Also read: Biomolecules

Lipids are a family of organic compounds, composed of fats and oils. These molecules yield high energy and are responsible for different functions within the human body. Listed below are some important characteristics of Lipids.
- Lipids are oily or greasy nonpolar molecules, stored in the adipose tissue of the body.
- Lipids are a heterogeneous group of compounds, mainly composed of hydrocarbon chains.
- Lipids are energy-rich organic molecules, which provide energy for different life processes.
- Lipids are a class of compounds characterised by their solubility in nonpolar solvents and insolubility in water.
- Lipids are significant in biological systems as they form a mechanical barrier dividing a cell from the external environment known as the cell membrane.
Also Read: Digestion and Absorption of Lipids
Lipids are the polymers of fatty acids that contain a long, non-polar hydrocarbon chain with a small polar region containing oxygen. The lipid structure is explained in the diagram below:

Lipid Structure – Saturated and Unsaturated Fatty Acids
Lipids can be classified into two main classes:
- Nonsaponifiable lipids
- Saponifiable lipids
Nonsaponifiable Lipids
A nonsaponifiable lipid cannot be disintegrated into smaller molecules through hydrolysis. Nonsaponifiable lipids include cholesterol, prostaglandins, etc
Saponifiable Lipids
A saponifiable lipid comprises one or more ester groups, enabling it to undergo hydrolysis in the presence of a base, acid, or enzymes , including waxes, triglycerides, sphingolipids and phospholipids.
Further, these categories can be divided into non-polar and polar lipids.
Nonpolar lipids, namely triglycerides, are utilized as fuel and to store energy.
Polar lipids, that could form a barrier with an external water environment, are utilized in membranes. Polar lipids comprise sphingolipids and glycerophospholipids.
Fatty acids are pivotal components of all these lipids.
Within these two major classes of lipids, there are numerous specific types of lipids, which are important to life, including fatty acids, triglycerides, glycerophospholipids, sphingolipids and steroids. These are broadly classified as simple lipids and complex lipids.
Also read: Biomolecules in Living Organisms
Simple Lipids
Esters of fatty acids with various alcohols.
- Fats: Esters of fatty acids with glycerol. Oils are fats in the liquid state
- Waxes : Esters of fatty acids with higher molecular weight monohydric alcohols
Complex Lipids
Esters of fatty acids containing groups in addition to alcohol and fatty acid.
- Phospholipids : These are lipids containing, in addition to fatty acids and alcohol, phosphate group. They frequently have nitrogen-containing bases and other substituents, eg, in glycerophospholipids the alcohol is glycerol and in sphingophospholipids the alcohol is sphingosine.
- Glycolipids (glycosphingolipids) : Lipids containing a fatty acid, sphingosine and carbohydrate.
- Other complex lipids : Lipids such as sulfolipids and amino lipids. Lipoproteins may also be placed in this category.
Precursor and Derived Lipids
These include fatty acids, glycerol, steroids, other alcohols, fatty aldehydes, and ketone bodies, hydrocarbons, lipid-soluble vitamins, and hormones. Because they are uncharged, acylglycerols (glycerides), cholesterol, and cholesteryl esters are termed neutral lipids. These compounds are produced by the hydrolysis of simple and complex lipids.
Some of the different types of lipids are described below in detail.
Fatty Acids
Fatty acids are carboxylic acids (or organic acid), usually with long aliphatic tails (long chains), either unsaturated or saturated.
- Saturated fatty acids
Lack of carbon-carbon double bonds indicate that the fatty acid is saturated. The saturated fatty acids have higher melting points compared to unsaturated acids of the corresponding size due to their ability to pack their molecules together thus leading to a straight rod-like shape.
- Unsaturated fatty acids
Unsaturated fatty acid is indicated when a fatty acid has more than one double bond.
“Often, naturally occurring fatty acids possesses an even number of carbon atoms and are unbranched.”
On the other hand, unsaturated fatty acids contain a cis-double bond(s) which create a structural kink that disables them to group their molecules in straight rod-like shape.
Role of Fats
Fats play several major roles in our body. Some of the important roles of fats are mentioned below:
- Fats in the correct amounts are necessary for the proper functioning of our body.
- Many fat-soluble vitamins need to be associated with fats in order to be effectively absorbed by the body.
- They also provide insulation to the body.
- They are an efficient way to store energy for longer periods.
Also Read: Fats
There are different types of lipids. Some examples of lipids include butter, ghee, vegetable oil, cheese, cholesterol and other steroids, waxes, phospholipids, and fat-soluble vitamins. All these compounds have similar features, i.e. insoluble in water and soluble in organic solvents, etc.
Waxes are “esters” (an organic compound made by replacing the hydrogen with acid by an alkyl or another organic group) formed from long-alcohols and long-chain carboxylic acids.
Waxes are found almost everywhere. The fruits and leaves of many plants possess waxy coatings, that can safeguard them from small predators and dehydration.
Fur of a few animals and the feathers of birds possess the same coatings serving as water repellants.
Carnauba wax is known for its water resistance and toughness (significant for car wax).
Phospholipids

Membranes are primarily composed of phospholipids that are Phosphoacylglycerols.
Triacylglycerols and phosphoacylglycerols are the same, but, the terminal OH group of the phosphoacylglycerol is esterified with phosphoric acid in place of fatty acid which results in the formation of phosphatidic acid.
The name phospholipid is derived from the fact that phosphoacylglycerols are lipids containing a phosphate group.
Our bodies possess chemical messengers known as hormones , which are basically organic compounds synthesized in glands and transported by the bloodstream to various tissues in order to trigger or hinder the desired process.
Steroids are a kind of hormone that is typically recognized by their tetracyclic skeleton, composed of three fused six-membered and one five-membered ring, as seen above. The four rings are assigned as A, B, C & D as observed in the shade blue, while the numbers in red indicate the carbons.
Cholesterol
- Cholesterol is a wax-like substance, found only in animal source foods. Triglycerides, LDL, HDL, VLDL are different types of cholesterol found in the blood cells.
- Cholesterol is an important lipid found in the cell membrane. It is a sterol, which means that cholesterol is a combination of steroid and alcohol. In the human body, cholesterol is synthesized in the liver.
- These compounds are biosynthesized by all living cells and are essential for the structural component of the cell membrane.
- In the cell membrane, the steroid ring structure of cholesterol provides a rigid hydrophobic structure that helps boost the rigidity of the cell membrane. Without cholesterol, the cell membrane would be too fluid.
- It is an important component of cell membranes and is also the basis for the synthesis of other steroids, including the sex hormones estradiol and testosterone, as well as other steroids such as cortisone and vitamin D.
Also Refer: Vitamins and Minerals
Frequently Asked Questions
What are lipids.
Lipids are organic compounds that are fatty acids or derivatives of fatty acids, which are insoluble in water but soluble in organic solvents. For eg., natural oil, steroid, waxes.
How are lipids important to our body?
Lipids play a very important role in our body. They are the structural component of the cell membrane. They help in providing energy and produce hormones in our body. They help in the proper digestion and absorption of food. They are a healthy part of our diet if taken in proper amounts. They also play an important role in signalling.
How are lipids digested?
The enzyme lipase breaks down fats into fatty acids and glycerol, which is facilitated by bile in the liver.
What is lipid emulsion?
It refers to an emulsion of lipid for human intravenous use. These are also referred to as intralipids which is the emulsion of soybean oil, glycerin and egg phospholipids. It is available in 10%, 20% and 30% concentrations.
How are lipids metabolized?
Lipid metabolism involves the oxidation of fatty acids to generate energy to synthesize new lipids from smaller molecules. The metabolism of lipids is associated with carbohydrate metabolism as the products of glucose are converted into lipids.
How are lipids released in the blood?
The medium-chain triglycerides with 8-12 carbons are digested and absorbed in the small intestine. Since lipids are insoluble in water, they are carried to the bloodstream by lipoproteins which are water-soluble and can carry the lipids internally.
What are the main types of lipids?
There are two major types of lipids- simple lipids and complex lipids. Simple lipids are esters of fatty acids with various alcohols. For eg., fats and waxes. On the contrary, complex lipids are esters of fatty acids with groups other than alcohol and fatty acids. For eg., phospholipids and sphingolipids.
What are lipids made up of?
Lipids are made up of a glycerol molecule attached to three fatty acid molecules. Such a lipid is called triglyceride.

Put your understanding of this concept to test by answering a few MCQs. Click ‘Start Quiz’ to begin!
Select the correct answer and click on the “Finish” button Check your score and answers at the end of the quiz
Visit BYJU’S for all Biology related queries and study materials
Your result is as below
Request OTP on Voice Call
Leave a Comment Cancel reply
Your Mobile number and Email id will not be published. Required fields are marked *
Post My Comment
Intriguing. I often wonder if high cholesterol isn’t mirroring high blood sugar, in that some form of cell resistance is at work.
No Doubt. It’s very very outstanding. Details I liked the most. I would like to thank all the people who published this lesson and to those who actually wrote this. It’s awesome. It builds a nice understanding to the students…I mean just like me…I hate the lipids since I was in ninth class….and now I am in the first year. And now after learning from this website…I think biology is my favourite subject and lipids is my favourite topic…Thanks a lot again to all of you.
I AGREE with you!
Very good apps, which I was looking for, found everything in it.
outstsanding. all one need about lipids is here
Wow it is wonderful I really appreciate the author of the write up. I look forward to know more
Thank you so much really helpful, appreciated.
This is really helping big thanks to the publishers
Wow, wow Thank U so much Really, I am so happy to help me with this.
I kindly needed help how to answer this question ‘How does the study of lipids start?’
Register with BYJU'S & Download Free PDFs
Register with byju's & watch live videos.

Microbe Notes
Lipids: Properties, Structure, Classification, Types, Functions
Lipids are a group of diverse macromolecules consisting of fatty acids and their derivatives that are insoluble in water but soluble in organic solvents.
- Lipids consist of fats , oils , hormones , and certain components of membranes that are grouped together because of their hydrophobic interactions.
- The lipids are essential constituents of the diet because of their high energy value.
- These are also essential for the fat-soluble vitamins and the essential fatty acids found with the fat of the natural foodstuffs.
- Fats combined with proteins (lipoproteins) are essential constituents of the cell membranes and mitochondria of the cell.
- Lipids occur naturally in living beings like plants, animals, and microorganisms that form various components like cell membranes, hormones, and energy storage molecules.
- Lipids exist in either liquid or non-crystalline solids at room temperatures and are colorless, odorless, and tasteless.
- These are composed of fatty acids and glycerol.
Table of Contents
Interesting Science Videos
Properties of Lipids
- Lipids may be either liquids or non-crystalline solids at room temperature.
- Pure fats and oils are colorless, odorless, and tasteless.
- They are energy-rich organic molecules
- Insoluble in water
- Soluble in organic solvents like alcohol, chloroform, acetone, benzene, etc.
- No ionic charges
- Solid triglycerols (Fats) have high proportions of saturated fatty acids.
- Liquid triglycerols (Oils) have high proportions of unsaturated fatty acids.
1. Hydrolysis of triglycerols
Triglycerols like any other esters react with water to form their carboxylic acid and alcohol– a process known as hydrolysis.
2. Saponification:
Triacylglycerols may be hydrolyzed by several procedures, the most common of which utilizes alkali or enzymes called lipases. Alkaline hydrolysis is termed saponification because one of the products of the hydrolysis is a soap, generally sodium or potassium salts of fatty acids.
3. Hydrogenation
The carbon-carbon double bonds in unsaturated fatty acids can be hydrogenated by reacting with hydrogen to produce saturated fatty acids.
4. Halogenation
Unsaturated fatty acids, whether they are free or combined as esters in fats and oils, react with halogens by addition at the double bond(s). The reaction results in the decolorization of the halogen solution.
5. Rancidity:
The term rancid is applied to any fat or oil that develops a disagreeable odor. Hydrolysis and oxidation reactions are responsible for causing rancidity. Oxidative rancidity occurs in triacylglycerols containing unsaturated fatty acids.
Structure of Lipids
- Lipids are made of the elements Carbon, Hydrogen and Oxygen, but have a much lower proportion of water than other molecules such as carbohydrates .
- Unlike polysaccharides and proteins, lipids are not polymers—they lack a repeating monomeric unit.
- They are made from two molecules: Glycerol and Fatty Acids.
- A glycerol molecule is made up of three carbon atoms with a hydroxyl group attached to it and hydrogen atoms occupying the remaining positions.
- Fatty acids consist of an acid group at one end of the molecule and a hydrocarbon chain, which is usually denoted by the letter ‘R’.
- They may be saturated or unsaturated .
- A fatty acid is saturated if every possible bond is made with a Hydrogen atom, such that there exist no C=C bonds.
- Unsaturated fatty acids, on the other hand, do contain C=C bonds. Monounsaturated fatty acids have one C=C bond, and polyunsaturated have more than one C=C bond.
Classification of Lipids
Lipids can be classified according to their hydrolysis products and according to similarities in their molecular structures. Three major subclasses are recognized:
1. Simple lipids
(a) Fats and oils which yield fatty acids and glycerol upon hydrolysis.
(b) Waxes , which yield fatty acids and long-chain alcohols upon hydrolysis.
Fats and Oils
- Both types of compounds are called triacylglycerols because they are esters composed of three fatty acids joined to glycerol, trihydroxy alcohol.
- The difference is on the basis of their physical states at room temperature. It is customary to call a lipid a fat if it is solid at 25°C, and oil if it is a liquid at the same temperature.
- These differences in melting points reflect differences in the degree of unsaturation of the constituent fatty acids.
- Wax is an ester of long-chain alcohol (usually mono-hydroxy) and a fatty acid.
- The acids and alcohols normally found in waxes have chains of the order of 12-34 carbon atoms in length.
2. Compound lipids
(a) Phospholipids , which yield fatty acids, glycerol, amino alcohol sphingosine, phosphoric acid and nitrogen-containing alcohol upon hydrolysis.
They may be glycerophospholipids or sphingophospholipid depending upon the alcohol group present (glycerol or sphingosine).
(b) Glycolipids , which yield fatty acids, sphingosine or glycerol, and a carbohydrate upon hydrolysis.
They may also be glyceroglycolipids or sphingoglycolipid depending upon the alcohol group present (glycerol or sphingosine).
3. Derived lipids:
Hydrolysis product of simple and compound lipids is called derived lipids. They include fatty acid, glycerol, sphingosine and steroid derivatives.
Steroid derivatives are phenanthrene structures that are quite different from lipids made up of fatty acids.
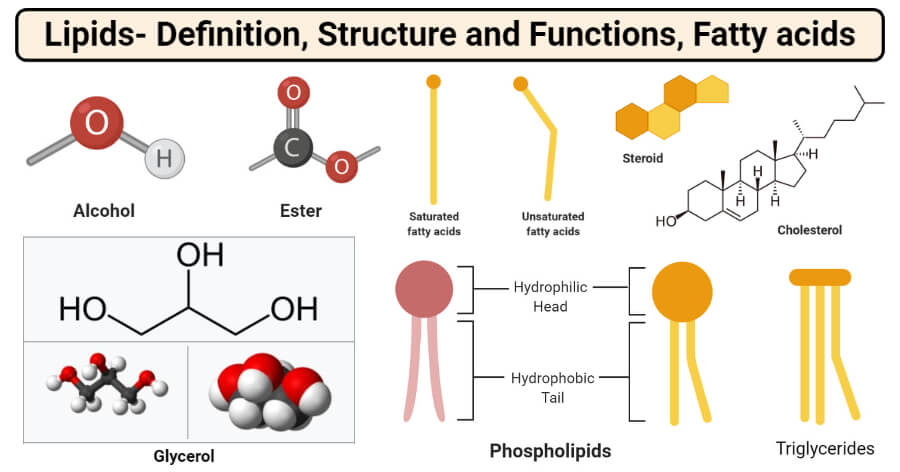
Alcohols and Esters
- The most important and frequently occurring alcohol found in lipids is glycerol. Glycerol is a small organic molecule consisting of three hydroxyls (OH-) groups.
- Glycerol makes up simple lipids which are esters of fatty acids and glycerol and similar alcohols.
- The alcohol might be glycerol or other long-chain alcohol. The long-chain alcohols are mostly mono-hydroxy with a single OH group.
- Depending on the alcohol used, simple lipids consist of fats, oil, or waxes. Fats and oils are esters of fatty acids and glycerol, whereas waxes are esters of fatty acids and long-chain alcohols.
- The esters of fatty acids are formed after the dehydration reaction between the fatty acids and the alcohol molecules.
Triglycerides
Triglycerides are a type of lipid which is an ester of three fatty acids with glycerol. Triglycerides are the main constituents of body fat in humans, other vertebrates, and vegetable fats.
Structure of Triglycerides
Triglycerides are tri-esters where three fatty acid molecules are bound to a single glycerol molecule by covalent ester bonds.
HOCH 2 CH(OH)CH 2 OH + RCO 2 H + R′CO 2 H + R″CO 2 H → RCO 2 CH 2 CH(O 2 CR′)CH 2 CO 2 R″ + 3H 2 O
- The three fatty acids involved in the condensation reaction are usually different, and their chain length also differs from one another.
- In naturally occurring triglycerides, the fatty acid chains mostly contain 16, 18, or 20 carbon atoms.
- Even-numbered carbon atoms present in animals and plants indicating the pathway of their biosynthesis from two-carbon acetyl CoA.
- Simple triglycerides might also have identical fatty acids forming homotriglycerides.
- The charges in triglycerides are evenly distributed around the molecules, which prevents the formation of hydrogen bonds with water molecules, making them insoluble in water.

Functions of Triglycerides
- Triglycerides are important macromolecules as they store most of the energy in the body.
- These are stored in fat cells which are then released into the bloodstream by the action of different hormones whenever necessary.
- The fat stored in the body forms a layer of insulation beneath the skin, which helps to maintain the body temperature.
- Triglycerides also aid in the absorption and transport of fat-soluble vitamins in the body.
What are Fatty acids?
- Fatty acids are organic molecules that are long-chained carboxylic acids with 4-36 carbon atoms.
- The hydrocarbon chains are either saturated or unsaturated, depending on the bonds between the carbon atoms. If all the carbon-carbon bonds are single, the acid is saturated; if one or more carbon-carbon double bonds are present, the acid is unsaturated.
- Naturally occurring fatty acids are mostly unbranched, and these occur in three main classes of lipids; triglycerides, phospholipids, and cholesteryl esters.
- Fatty acids are not found in the free state but remain associated with alcohol to form triglycerides.
- Fatty acids are stored as an energy reserve (fat) through an ester linkage to glycerol to form triglycerides.
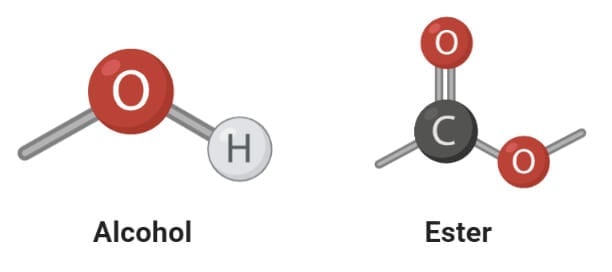
Saturated and Unsaturated Fatty acids
1. saturated fatty acids.
- Saturated fatty acids are the simplest form of fats that are unbranched linear chains of CH 2 groups linked together by carbon-carbon single bonds with a terminal carboxylic acid.
- The term ‘saturated’ is used to indicate that the maximum number of hydrogen atoms are bonded to each carbon atom in a molecule of fat.
- The general formula for these acids is C n H2 n +1COOH.
- Fatty acids obtained from an animal source are mostly even-numbered linear chains of saturated fatty acids.
- Saturated fatty acids usually have a higher melting point than their counterparts which is why saturated fatty acids remain in the solid-state at room temperatures.
- These are mostly solid and are found in animal fat like butter, meat, and whole milk. But some saturated fatty acids are also found in vegetable sources like vegetable oil, coconut oil, and peanut oil.
2. Unsaturated fatty acids
- Unsaturated fatty acids are more complex fatty acids with bent hydrocarbon chains linked together by one or more carbon-carbon double bonds with a terminal carboxylic acids group.
- The term ‘unsaturated’ indicates that the carbons atoms do not have the maximum possible hydrogen atoms bound to carbon atoms.
- Due to the presence of double bonds, the cis and trans conformation of these molecules are important. The unsaturated fatty acids found in the human body exist in the cis conformation
- Unsaturated fatty acids have a lower melting point as compared to saturated fatty acids, and thus they exist in the liquid state at room temperatures
- Most vegetable oils and fish oils are some of the important sources of unsaturated fatty acids.
Read Also: 20 Differences Between Saturated and Unsaturated fatty acids
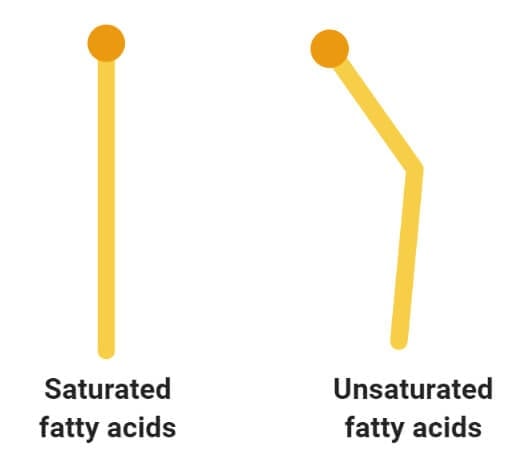
Glycerol and the formation of ester bonds.
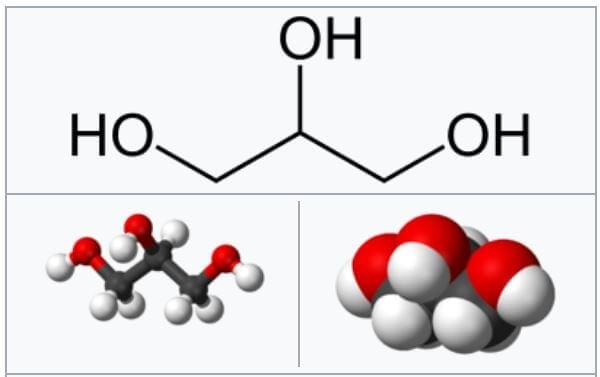
Image Source: Wikipedia .
- Glycerol is a simple organic compound in three hydroxyl groups that is a colorless, odorless, and viscous liquid.
- It forms the backbone of many lipids that are termed glycerides. The fat is later hydrolyzed into fatty acids and glycerol where the fatty acid provides energy to the body, whereas the glycerol is converted into glucose.
- The reaction involved in the formation of ester bonds is termed as condensation reaction where the free hydroxyl end of the glycerol molecule joins to the OH of the COOH group of the fatty acid.
- The process of condensation is termed esterification due to the formation of ester bonds between the two molecules.
- The lipid molecules formed from three fatty acids and a single glycerol molecule are termed as triacylglycerols or triglycerides.
Phospholipids
- A phospholipid is an organic molecule consisting of fatty acids, a phosphate group, and a glycerol group that forms the main component of various cellular membranes.
- Phospholipid bilayer forms an important part of the cell membrane for the selective transport of molecules in and out of the cell.
- The phosphate group forms the hydrophilic head, whereas the fatty acids form the hydrophobic tails. The head and tail regions in phospholipids are joined by a glycerol molecule.
- The hydrophobic and hydrophilic interaction between different molecules and the lipid bilayer enables the passage of biomolecules. These interactions make the cell membrane amphipathic.
1. Hydrophilic (polar) phosphate heads
- The hydrophilic head or water-loving part of the phospholipids contains a negatively charged phosphate group with an unidentified alkyl group.
- The hydrophilic region might or might not be polar or charged.
- The heads of the phospholipid membrane face outwards that remain in interaction with the aqueous solution inside and outside the cell.
- As water is a polar molecule, the hydrophilic head immediately forms electrostatic interaction with the water molecule.
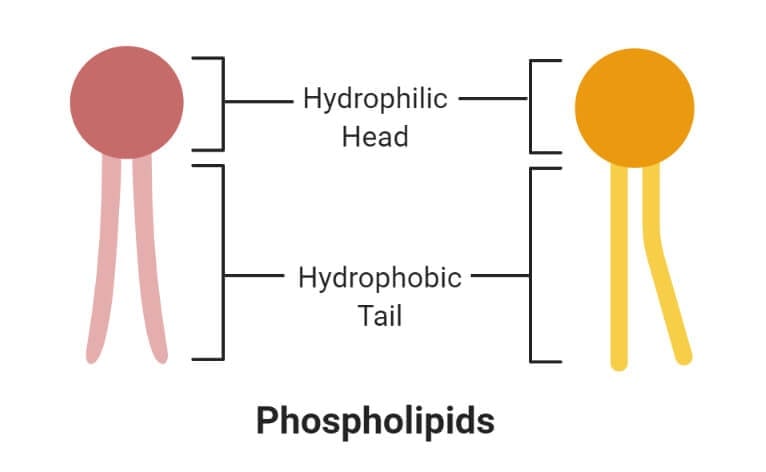
2. Hydrophobic (non-polar) fatty acid tails
- The hydrophobic part of the phospholipid bilayer is also termed the water-fearing portion that consists of long non-polar fatty acid tails.
- These tails easily interact with other hydrophobic molecules but do not interact with water molecules.
- The tail region is a non-polar end where charge-less molecules are present.
- The hydrophobic tails are thus tucked towards the interior of the membrane in order to shield the tails from the surrounding water. This arrangement is also energetically favorable.
- The hydrophobic interactions form a good barrier between the inside and outside of the cell as water, and other charge molecules cannot easily cross the hydrophobic core of the membrane.
Sterols (Cholesterol)
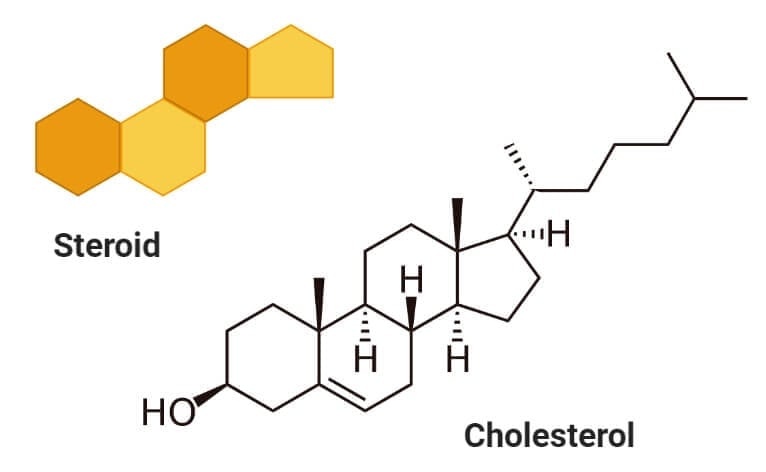
- Sterols are a type of lipids composed of steroid alcohols occurring naturally in plants, animals, fungi, and several bacteria.
- The most important and familiar type of sterol is cholesterol which plays an essential role in cell membrane structure and functions.
- Cholesterol acts as a precursor to fat-soluble vitamins like Vitamin D and hormones.
- Cholesterol is formed of four linked hydrocarbon rings forming the bulk of the steroid structure. One end of cholesterol consists of a hydrocarbon tail, whereas the other end is linked to an alcohol group.
- The hydroxyl group joins with other hydroxyl groups or carbonyl oxygen of phospholipids.
- Cholesterol can be biosynthesized within the body of various animals. In humans, the liver makes up 100% of all cholesterol required for the body.
- Cholesterol is considered essential for the regulation of membrane fluidity in animals. It also increases the permeability of the cell membrane to sodium and potassium ions.
- However, if the concentration of cholesterol increases beyond normal, it might combine with other components in the blood and form plaque. The plaque might attach to the walls of arteries and veins, resulting in coronary artery disease.
Functions of Lipids
- Biological lipids are a chemically diverse group of compounds, and the biological functions of the lipids are as diverse as their chemistry.
- In the body, fats serve as an efficient source of energy and are also stored in the adipose tissues. These also serve as an insulating material in the subcutaneous tissues and around certain organs.
- Phospholipids and sterols are major structural elements of biological membranes.
- Similarly, fats combined with proteins (lipoproteins) are important constituents of the cell membranes and mitochondria of the cell.
- Lipids also act as the structural component of the cell and provide the hydrophobic barrier that allows the separation of the aqueous contents of the cell and subcellular structures.
- Other lipids, although present in relatively small quantities, play crucial roles as enzyme cofactors, electron carriers, light-absorbing pigments, and hydrophobic anchors for proteins.
- Lipids are also activators of enzymes like glucose-6-phosphatase, β-hydroxybutyric dehydrogenase, and stearyl CoA desaturase.
References and Sources
- Jain JL, Jain S, and Jain N (2005). Fundamentals of Biochemistry. S. Chand and Company.
- Nelson DL and Cox MM. Lehninger Principles of Biochemistry. Fourth Edition.
- Berg JM et al. (2012) Biochemistry. Seventh Edition. W. H Freeman and Company.
- Biologydictionary.net Editors. (2016, November 08). Phospholipid. Retrieved from https://biologydictionary.net/phospholipid/
- Smith, C. M., Marks, A. D., Lieberman, M. A., Marks, D. B., & Marks, D. B. (2005). Marks’ basic medical biochemistry: A clinical approach. Philadelphia: Lippincott Williams & Wilkins.
- 3% – http://ndl.ethernet.edu.et/bitstream/123456789/78706/12/Chap-12.pdf
- 2% – http://fac.ksu.edu.sa/sites/default/files/4-bch302_lipids_i_0.pdf
- 1% – https://www.britannica.com/science/lipid
- 1% – https://noahstrength.com/health/three-kinds-of-triglycerides/
- 1% – https://letslearnplants.blogspot.com/
- 1% – https://ibiologia.com/phospholipid-bilayer/
- 1% – https://hyperleap.com/topic/Cholesterol
- 1% – https://chem.libretexts.org/Bookshelves/Organic_Chemistry/Map%3A_Organic_Chemistry_(McMurry)/27%3A_Biomolecules_-_Lipids/27.03%3A_Waxes_Fats_and_Oils
- 1% – https://biologydictionary.net/hydrophilic/
- 1% – http://www.bioinfo.org.cn/book/biochemistry/chapt09/sim1.htm
About Author
Anupama Sapkota
3 thoughts on “Lipids: Properties, Structure, Classification, Types, Functions”
These notes come in handy. Really really appreciate this effort of combining all these. Thankyou. Very informative material, and such precise and to the point written.
This is astounding introductory notes for lipids under biochemistry, I really enjoyed the part where the concepts of lipid biomolecule connected to chemistry bonds and other stuff like the condensation
Leave a Comment Cancel reply
Save my name, email, and website in this browser for the next time I comment.
This site uses Akismet to reduce spam. Learn how your comment data is processed .
Want to create or adapt books like this? Learn more about how Pressbooks supports open publishing practices.
Introduction to Lipids
Let’s take a closer look at the role of lipids now…
Functions of Lipids
The key functions of lipids in biological systems include:
- Energy Storage
- Water Barrier/Protection
- Cell Membranes
Lipids can be categorized into (3) functional groups:
Oils, fats, waxes.
- Energy storage, protection
- Phospholipids
- Construction of cell membranes
Lipid Function
Lipids are water-insoluble molecules which can be categorized into several groups.
- Oils, fats and waxes
- Energy storage (fats, oils)
- Protection from the environment (waxes)
- Forming cellular membranes (phospholipids)
- Signaling hormones (steroids)
BIO109 Biology I Introduction to Biology Copyright © by Diane Rinehart. All Rights Reserved.
Share This Book
Module 3: Important Biological Macromolecules
Introduction to lipids, illustrate different types of lipids and relate their structure to their role in biological systems.
In this outcome, we will discuss lipids, or fats, and the role they play in our bodies.
What You’ll Learn to Do
- Distinguish between the different kinds of lipids
- Identify several major functions of lipids
Learning Activities
The learning activities for this section include the following:
- Self Check: Lipids
- Introduction to Lipids. Authored by : Shelli Carter and Lumen Learning. Provided by : Lumen Learning. License : CC BY: Attribution

- Open access
- Published: 08 November 2024
Association analysis of gut microbiota with LDL-C metabolism and microbial pathogenicity in colorectal cancer patients
- Mingjian Qin 1 na1 ,
- Zigui Huang 1 na1 ,
- Yongqi Huang 1 na1 ,
- Xiaoliang Huang 1 ,
- Chuanbin Chen 1 ,
- Yongzhi Wu 1 ,
- Zhen Wang 1 ,
- Fuhai He 1 ,
- Binzhe Tang 1 ,
- Chenyan Long 1 ,
- Xianwei Mo 1 ,
- Jungang Liu 1 &
- Weizhong Tang 1
Lipids in Health and Disease volume 23 , Article number: 367 ( 2024 ) Cite this article
Metrics details
Colorectal cancer (CRC) is the most common gastrointestinal malignancy worldwide, with obesity-induced lipid metabolism disorders playing a crucial role in its progression. A complex connection exists between gut microbiota and the development of intestinal tumors through the microbiota metabolite pathway. Metabolic disorders frequently alter the gut microbiome, impairing immune and cellular functions and hastening cancer progression.
This study thoroughly examined the gut microbiota through 16S rRNA sequencing of fecal samples from 181 CRC patients, integrating preoperative Low-density lipoprotein cholesterol (LDL-C) levels and RNA sequencing data. The study includes a comparison of microbial diversity, differential microbiological analysis, exploration of the associations between microbiota, tumor microenvironment immune cells, and immune genes, enrichment analysis of potential biological functions of microbe-related host genes, and the prediction of LDL-C status through microorganisms.
The analysis revealed that differences in α and β diversity indices of intestinal microbiota in CRC patients were not statistically significant across different LDL-C metabolic states. Patients exhibited varying LDL-C metabolic conditions, leading to a bifurcation of their gut microbiota into two distinct clusters. Patients with LDL-C metabolic irregularities had higher concentrations of twelve gut microbiota, which were linked to various immune cells and immune-related genes, influencing tumor immunity. Under normal LDL-C metabolic conditions, the protective microorganism Anaerostipes_caccae was significantly negatively correlated with the GO Biological Process pathway involved in the negative regulation of the unfolded protein response in the endoplasmic reticulum. Both XGBoost and MLP models, developed using differential gut microbiota, could forecast LDL-C levels in CRC patients biologically.
Conclusions
The intestinal microbiota in CRC patients influences the LDL-C metabolic status. With elevated LDL-C levels, gut microbiota can regulate the function of immune cells and gene expression within the tumor microenvironment, affecting cancer-related pathways and promoting CRC progression. LDL-C and its associated gut microbiota could provide non-invasive markers for clinical evaluation and treatment of CRC patients.
Introduction
According to GLOBOCAN 2020 data from the International Agency for Research on Cancer (IARC), colorectal cancer (CRC) incidence ranks third, following breast and lung cancers, while its mortality rate is second only to lung cancer. Each year, over 1.9 million new CRC cases are predicted globally, with 0.935 million deaths [ 1 ]. Social and economic development, coupled with sedentary lifestyles and increased consumption of animal-derived foods, leads to reduced physical activity and obesity, which are independently related to CRC risk [ 2 ]. Although the mechanisms underlying CRC are unclear, research shows that obesity and sedentary lifestyles may cause lipid metabolism disorders [ 3 , 4 ]. Additionally, lipid metabolism disorders are increasingly recognized as important roles in cancer progression, including CRC [ 5 ]. Cholesterol, an important component of blood lipids, is highly lipophilic and is transported by lipoproteins, a lipid-protein complex. Low-density lipoprotein cholesterol (LDL-C), a primary type of lipoprotein, transports cholesterol from the liver to various tissues, providing raw materials for tissue cells, including cancer cells [ 6 ]. Studies have shown that LDL-C receptor levels are upregulated in CRC patients, and the addition of LDL-C to cell cultures significantly increases ROS levels in CRC cells, alters gene expression, and activates the MAPK pathway, thereby enhancing intestinal tumorigenicity and accelerating tumor progression. [ 7 ]. Moreover LDL-C mediates the occurrence of CRC through its oxidation to oxidized low-density lipoprotein (oxLDL) [ 8 ]. A similar study revealed that the interaction between LDL-C and the mucin family gene MUC4 rs1104760A > G may be important in diagnosing CRC. This combination may induce CRC by affecting LDL-C levels [ 9 ]. Meanwhile, retrospective cross-sectional studies have shown that elevated LDL-C levels are significantly associated with lymph node metastasis in various cancers, including CRC [ 10 ].
Primary prevention is crucial in reducing the global burden of CRC. Although endoscopic examinations can reduce CRC incidence and mortality, flexible sigmoidoscopy is ineffective for proximal colon cancer and challenging for large-scale screening due to cost and invasiveness [ 11 ]. With deeper understanding of CRC metagenomics, gut microbiota offer new perspectives for CRC diagnosis and therapy. Bacteria such as Parvimonas mira and Solobacterium moorei serve as non-invasive biomarkers for CRC [ 12 , 13 ], whereas Bacteroides vulgatus and Akkermansia muciniphila exhibit anti-cancer effects on CRC cell proliferation [ 14 ]. Additionally, gut microbiota can regulate host metabolism and show promise in studies on lipid metabolism associated with intestinal tumors. For example, P anaerobius enriched in colon tumors and adenoma tissues may interact with toll-like receptors to increase intracellular active oxidants, promoting cholesterol synthesis and CRC cell proliferation [ 15 ]. Squalene epoxidase(SQLE), an essential enzyme in cholesterol synthesis, can mediate intestinal tumor occurrence through the gut microbiota-metabolic axis [ 16 ].
Thus, LDL-C, a major type of cholesterol, has a close relationship with CRC and interacts with gut microbiota. While research has shown that intestinal microbiota preparations can lower human LDL-C levels [ 17 ], no studies have yet suggested a relationship between intestinal microbiota of CRC patients and LDL-C. This study sought to investigate the makeup and abundance of intestinal microbiota in the feces of CRC patients, identify internal relationships among typical gut microbiota and their relationships with elevated LDL-C levels, investigate microbial factors responsible for LDL-C metabolism disorders in CRC patients, and explore possible internal regulation among these gut microorganisms. Subsequent research will focus on the immune and biological mechanisms driven by typical gut microbiota in CRC development amidst irregular LDL-C metabolism, and create predictive models to biologically assess LDL-C levels in CRC patients.
Participant details and inclusion criteria
The Medical Ethics Committee of the Guangxi Medical University Cancer Hospital has approved this research protocol. All participants signed an informed consent form prior to surgery and were notified about the sampling before sample collection. Based on the inclusion criteria, researchers collected fecal samples collected from 236 CRC patients prior to treatment between January 1, 2021 and December 31, 2021, and ultimately collected 198 fecal samples that passed the 16S ribosomal RNA (16S rRNA) sequencing quality test. Concurrently, freshly collected tissue samples of the aforementioned subjects who underwent surgical treatment at the Guangxi Medical University Cancer Hospital were collected and stored in cryogenic liquid nitrogen. Among them, 181 cancer patients had LDL-C information. Additionally, among the 17 CRC patients who underwent transcriptomic sequencing of tumor tissue samples, LDL-C data were available for 14 samples, and 8 samples simultaneously underwent 16S rRNA sequencing from pre-treatment fecal samples.
The inclusion criteria for this study include: 1. Patients who underwent surgery and have a clear pathological classification (staging in accordance with the ACJJ CRC classification guidelines), or CRC patients diagnosed by colonoscopy histopathological biopsy; 2. No history of comorbidities or other malignant tumors in the past; 3. Excluding other gastrointestinal disorders, there are no acute complications such as complete bowel obstruction and gastrointestinal perforation; 4. Prior to collecting fecal specimens, none of the patients had received any cancer therapy, including surgical procedure, chemo, radiation therapy, immune therapy, and traditional Chinese medicine treatment; 5. Not using antibiotics and gut microbiota preparations within the past month; 6. unconscious disorders or other cognitive impairments.
Collection of stool samples and 16S rRNA sequencing
After receiving notification of the sampling plan, the subjects collected fecal samples on the second day of admission. During the sampling process, members of research group guided the subjects to avoid urine contamination, and used sterile fecal collection tubes to retain the middle part of the fecal sample. Subsequently, the fecal sample was stored in a sterile ice container, encased in a 2 mL EP tube with a dosage of 200 mg per tube, and preserved in a refrigerator at -80° C. Following the dispatch of fecal specimens to the lab with the MOBIO PowerSoil® DNA Isolation Kit, DNA was isolated from 200 mg of feces using Tris–EDTA buffer, adhering to the prescribed product guidelines. Following the extraction of DNA, the specimens undergo DNA quality testing, permitting those of satisfactory quality to advance to the subsequent experiment. Primers 341F (5′-CCTACGGGNGGCWGCAG-3′) and 805R (5′-GACTACHVGGGTATCTAATCC-3′) were used to focus on and secure the V3 and V4 segments of the 16S rRNA gene, followed by PCR amplification of these targeted sequences. Post-PCR amplification, the initial analysis of each sample’s PCR products was conducted through 2% agarose gel electrophoresis, aiming for a band size between 300 and 350 base pairs, with a sequencing depth of 50,000 reads to capture the target sequences. Subsequently, the PCR outputs were measured with the Quant-iT PicoGreen dsDNA Assay Kit, and all specimens were merged in equal molar amounts, adhering to the sequencing criteria derived from each sample’s quantitative outcomes. Subsequently, the KAPA Library Quantification Kit KK4824 was employed to measure the quantity of the mixed libraries. Ultimately, sequencing of the libraries was conducted using an Illumina PE250 device by Genesky Biotechnologies company, (Shanghai, China), employing a 2 × 250 bp approach after successful completion of the library preparation.
Tissue sample collection and transcriptome high throughput sequencing
Based on the premise that the interval between separation and storage in liquid nitrogen is within 30 min, fresh tissues, with soybean size, were obtained from surgically removed tumors and adjacent normal tissue. Using Trizol ® The Total RNA Extraction Kit extracted total RNA from 17 CRC tumor samples and detected the integrity of the RNA using electrophoresis. RNA purity was determined through micro ultraviolet spectrophotometers. Refer to the instructions of the RNA seq sample preparation kit (VAHTS ™ Stranded mRNA-seq Library Prep Kit for Illumina ®), remove rRNA and construct cDNA library. The transcriptome library sequencing was performed using the Illumina NovaSeq 6000 by GENE + company, (Beijing, China). The unprocessed sequencing dataset was evaluated for quality by FastQC, and the valid dataset was first compared with the reference genome using HISAT2 (version: hg38). Gene expression was evaluated using StringTie and known gene models, and the calculated TPM (Transcripts Per Million) values were used to quantify the expression abundance of each gene.
Analysis of tumor immune infiltration
Using the ‘CIBERSORT R script v1.03’ through R software, the CIBERSORT algorithm constructs a feature matrix derived from microarray data from tumor tissue sequencing. Subsequently, the TPM matrix was transformed into a relative abundance gene feature matrix of 22 immune cells (including B cells, CD4 + T cells, CD8 + T cells, neutrophils, macrophages, dendritic cells and various varieties and functional statuses of immune cells) for tumor immune infiltration analysis [ 18 ].
Functional annotation analysis of transcriptome sequencing related to LDL-C
The Single Sample Gene Set Enrichment Analysis (ssGSEA) [ 19 ] algorithm calculates the matrix of gene set scores for each sample using the GSVA software package v1.46.0, based on downloaded gmt format gene set files (c2.cp.kegg.v2022.1.Hs.symbols.gmt, c5.go.v2022.1.Hs.symbols.gmt). Next, the L-LDL-C group was used as the control group, the variations in Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways among the groups were examined using the limma algorithm in the TCGAbiolinks package v2.25.3. GO enrichment analysis encompasses three aspects: biological processes (BP), cellular components (CC) and molecular functions (MF). The threshold for statistical significance of differentially expressed genes is: P < 0.05 and |log2FC|> 0.
Construction and recognition of machine learning models for gut microbiome biomarkers
Using the multilayer perceptron (MLP) model and the XGBoost (XGB) model to identify gut microbiota markers, respectively, to predict LDL-C levels in CRC patients. MLP is a feedforward artificial neural network model, which comprises an input layer, several hidden layers, and an output layer. Employing backpropagation technology, MLP iteratively adjusts the weights between neurons, ultimately achieving the construction of a neural network between the input and output layers [ 20 ]. XGB, a boosting-based integration algorithm, uses information from previous trees to enhance the quality of the current tree for iterative generation by building learners in parallel [ 21 ]. As a typical integration of classification and regression tree cart algorithms, XGB has improved the traditional Gradient Boosting Decision Tree [ 22 ]. These improvements include the introduction of additional regularization, integrated tree pruning, and subsampling features in XGB, which significantly alleviate overfitting problems, as well as the use of techniques for calculating generalized gain scores to simplify optimization problems in boosting trees [ 23 ].
Linking the SciKit Learn 0.18( https://scikit-learn.org/stable/ ) Platform and Python, utilized downloaded installation packages to construct and assess machine learning models. According to a 7:3 ratio, microbiota dataset of 181 patients who met the inclusion criteria was randomly split into training and testing sets. Subsequently, MLP and XBG models were developed and predicted using LDL-C related differential gut microbiota species with differential importance in the top 15%. Finally, the receiver operating curve (ROC) and area under curve (AUC) were applied to assess the models’ accuracy performance.
Analysis method for 16S rRNA sequencing
Qualitative Insights Into Microbiological Ecology version 2 was used to perform quality filtering on the FASTQ raw sequencing data of all samples. Subsequently, species were annotated according to the Greengenes database v13.8, while intestinal microbiota ASV/OTU was extracted using the photoseq package v126.1. Firstly, the gut microbiota diversity within the group was evaluated employing α-diversity, where Chao1 and ACE characterized the species abundance of the microbiota, while Shannon and Simpson described the microbial diversity and evenness. Secondly, β-diversity was used to evaluate the variability of the microbial structure in each sample across distinct groups. ANOSIM and ADONIS analyses were performed employing the vegan package v2.5.6. Subsequently, the mixOmics v6.6.2 software package was employed to complete partial least squares discriminant analysis (PLS-DA). Next, Linear discriminant analysis Effect Size (LEfSe) analysis was performed employing LEfSe software v1.0.0, combined with linear discriminant analysis (LDA) to evaluate analysis results, in order to identify species with significant abundance differences between groups (employing |LDA|> 2 and P < 0.05 as difference screening thresholds). Ultimately, employing PICRUSt2 software 2.3.0 to predict the KEGG pathways enriched between sequencing sample groups, and calling vegan package v2.5.6, the study used non-parametric Mann–Whitney U rank-sum test to analyze the inter group diversity indices and KEGG pathway variability. Finally, the ggplot2 package v3.4.0 was used to visualize histograms. The above operations were all completed using R software v3.5.1.
Statistical methods
Using SPSS software v23.0, continuous data analysis was performed on clinical data using t-tests, while quantitative data analysis was performed using Pearson Chi-square test. The subsequent procedures were completed using R software v4.2.2. Pearson correlation analysis was used to measure the correlation between gut microbiota and immune cell abundance with immune-related genes. The ggcorplot package v0.1.4 was used to perform Spearman correlation analysis to evaluate the correlation between different subgroups of differential gut microbiota, the correlation between differential microbiota and KEGG pathway, and the correlation between intergroup differential gut microbiota and BP and MF projects. Among them, the ggcorplot software package v0.1.4, Igraph software package v1.3.5, and Cytoscope software v3.7.2 were used to visualize the relevant matrices.
Essential information and clinical features of CRC patients classified by LDL-C levels
Following the application of inclusion and exclusion criteria, patients possessing pre-treatment LDL-C information were enrolled and divided into H-LDL-C and L-LDL-C groups according to their preoperative LDL-C levels. The H-LDL-C group included 80 CRC patients with LDL-C values above 3.37 mmol/L(129.62 mg/dL), while the L-LDL-C group included 101 CRC patients whose LDL-C levels at or underneath the maximum threshold of normal values (the reference range for normal values is 0–3.37 mmol/L (129.62 mg/dL)). As Table 1 shows, CRC patients with different LDL-C levels did not differ significantly in age or sex, suggesting balanced and comparable baseline data. Differences in serum triglyceride levels, serum albumin levels, and Body Mass Index were not statistically significant, indicating comparable nutritional status between the two groups. Additionally, patients with H-LDL-C had a higher percentage of abnormal total cholesterol compared to those in the L-LDL-C group ( P < 0.001), while high-density lipoprotein cholesterol (HDL-C) did not differ significantly between the two groups, suggesting that LDL-C might be associated with cholesterol metabolism disorders in CRC patients.
Comparison of microbial diversity between H-LDL-C and L-LDL-C groups in CRC Patients
At the start of the study, differences in microbial diversity between the H-LDL-C and L-LDL-C groups of CRC patients were investigated using α-diversity and β-diversity indices. α-diversity for samples from both patient groups is shown in Fig. 1 A. Although differences were observed, they were not statistically significant ( P > 0.05). Figure 1 B presents the β-diversity for two CRC patient groups. Bray ( P = 0.3107) and Jaccard ( P = 0.2659) indices suggest no statistically significant differences in gut microbiota composition between the groups (Supplementary Tables 1 and 2). PLS-DA analysis, shown in Fig. 1 C, revealed that CRC patients in two groups clustered according to their gut microbiota. The findings suggest that although there were no statistically significant differences in fecal microbiota diversity within and between groups, CRC patients persist substantial differences in gut microbiota composition based on LDL-C levels.
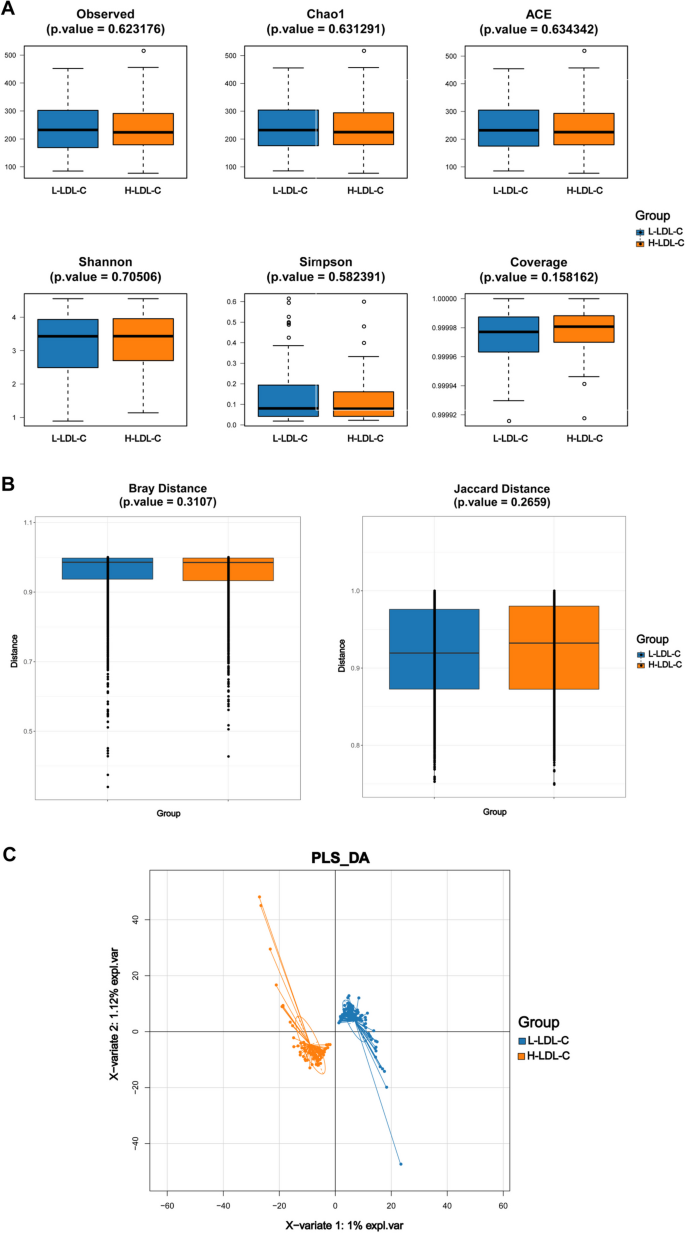
Comparison of gut microbiota diversity index between L-LDL-C group and H-LDL-C group patients with CRC. A Comparison of α-diversity index of gut microbiota between L-LDL-C group and H-LDL-C group in CRC patients. B Comparison of β-diversity index of gut microbiota between L-LDL-C group and H-LDL-C group in CRC patients. The horizontal axis represents the group, the vertical axis represents the diversity index value of the sample community within the group, and the color also represents the group. C PLS-DA analysis of gut microbiota in the L-LDL-C and H-LDL-C groups of CRC patients. The dots represent each sample of gut microbiota, the color represents the group, the horizontal and vertical axis scales represent the relative distance of each sample, and X variable 1 and X variable 2 represent the factors that affect the changes in gut microbiota composition of CRC patients in the L-LDL-C and H-LDL-C groups, respectively
Identification of gut microbiota associated with abnormal LDL-C metabolism
To investigate gut microbiota with varying abundance between H-LDL-C and L-LDL-C groups and identify key biomarkers for abnormal LDL-C metabolism, the study performed LEfSe analysis on these two groups. The analysis revealed significant statistical variations in the abundance of 24 microbial communities between two groups. Both H-LDL-C and L-LDL-C groups exhibited significantly greater abundance of 12 microbial communities compared to the other group ( P < 0.05; see Supplementary Table 3; Fig. 2 A, B). Figure 2 B displays LDA scores for top10 in each group of these 24 differential microbial communities after LEfSe analysis (log10 transformed). Higher scores indicate greater significance for these species. Correlations between dominant microbial communities in two groups were plotted (Fig. 2 C) to further explore their relationships with LDL-C. Among these, four dominant microbial communities in the L-LDL-C group, f__Veillonellaceae.g__Veillonella , f__Corynebacteriaceae.g__Corynebacterium , f__Bifidobacteriaceae.g__S-cardovia and o__Ac-tinomycetales. f__Corynebacteriaceae , were most closely associated with other nodes. These indicate that these four dominant microbial communities are closely related to other dominant microbial communities. Simultaneously, f__Veillonellaceae.g__Veillonella , a dominant microbial community in the L-LDL-C group showed a negative correlation with f__Coriobacteriaceae.g__Paraeg-gerthella , g__Fusobacterium.s__Fusobacterium_necrophorum, and g__Coprobacillus.s__uncultured_organism in the H-LDL-C group. These results indicate that negative regulatory interactions may occur among the dominant microbial communities.

Analysis of differences in gut microbiota between L-LDL-C group and H-LDL-C group CRC patients. A Evolutionary relationship diagram of LEfSe analysis. The node size represents the species abundance and is directly proportional to the species abundance. Node color represents grouping, and yellow nodes in branches represent species with no significant differences in abundance between groups; the red nodes represent species with significantly higher abundance in the L-LDL-C group, while the green nodes represent species with significantly higher abundance in the H-LDL-C group. Each layer node represents a phylum/class/order/family/genus/species from the inside out, and the annotations for each layer's species markers represent a phylum/class/order/family/genus/species from the outside in. B LDA bar chart based on 16S rRNA gene sequencing. The color of the bar chart represents the group, the horizontal coordinate represents the LDA score (after log10 processing), the vertical coordinate represents the species with significantly higher abundance in the group, and the length of the bar chart represents the size of the LDA score value. C LDL-C related differences in gut microbiota correlation network diagram. Each node represents each species, node color represents group, node size represents the number of edges connected to the node. The larger the node, the more edges connected to the node. The connecting line indicates a significant correlation between the two nodes. The blue line represents Spearman correlation coefficient values below 0 (negative correlation), while Spearman correlation coefficient values above 0 (positive correlation) are represented by the red line. The thicker the red line, the greater the Spearman correlation coefficient between two nodes
Predicting gut microbiota function in H-LDL-C and L-LDL-C groups
Next, PICRUSt2 (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States 2) software was used to analyze enriched KEGG pathways among characteristic microbiota in two groups to explore the biological relationship between LDL-C and its related gut microbiota. Among the 180 KEGG pathways analyzed, four pathways exhibited statistically significant differences ( P < 0.05). The Hypertrophic Cardiomyopathy pathway ( P = 0.047) was abundant in H-LDL-C group, while the Steroid hormone biosynthesis ( P = 0.042), Steroid biosynthesis ( P = 0.042), and Biosynthesis of siderophore group nonribosomal peptides ( P = 0.0499) were significantly more abundant in the L-LDL-C group than in the H-LDL-C group (Supplementary Fig. 1 and Supplementary Table 4). These results suggest that gut microbiota associated with LDL-C are closely linked to lipid metabolism in CRC patients.
Relationship between differential gut microbiota associated with LDL-C and immune cells
Tumor-infiltrating immune cells are those that enter the tumor microenvironment (TME) and interact with it, playing a role in either promoting or inhibiting tumor growth. To investigate the connection between LDL-C-associated intestinal microbiota and tumor-infiltrating immune cells, the study created a bar chart to show the composition of 22 immune cells from 14 CRC patients with LDL-C and RNA sequencing dataset (Fig. 3 A). Figure 3 A shows that each CRC patient has a unique immune cell composition in the TME. Overall, the H-LDL-C group had high proportions of follicular helper T cells (Tfh) and regulatory T cells (Tregs). Conversely, the L-LDL-C group had a high proportion of plasma cells.

The correlation between LDL-C related gut microbiota and tumor immune infiltrating cells. A Bar chart of relative abundance of immune cells in CRC patients grouped by LDL-C status. Each bar represents a sample, and the vertical coordinates represent the predicted relative abundance values of immune cells. The sum of the relative abundances of all immune cells in a single sample is 1, and each color in the graph corresponds to one type of immune cell. B Heat map of the correlation between dominant microbial communities and immune cell abundance in the H-LDL-C group. C Heat map of the correlation between dominant microbial communities and immune cell abundance in the L-LDL-C group. The horizontal axis represents immune cells, and the vertical axis represents microbiota. In the figure, red represents positive correlation, blue represents negative correlation, color depth represents the magnitude of Pearson correlation coefficient, and color from light to dark represents the value of Pearson correlation coefficient from small to large. The “*” in the figure represents the size of the P -value: none * represents a P -value ≥ 0.05, * represents 0.01 ≤ P < 0.05, * * represents 0.001 ≤ P < 0.01, and * * * represents P < 0.001. D Network diagram showing the correlation between LDL-C related differential gut microbiota and immune cells. Each node represents each gut microbiota or immune cell and the connecting line represents a significant correlation between the two nodes; the blue line indicates that the Pearson correlation coefficient is less than 0 (negative correlation), while the red line indicates that the Pearson correlation coefficient is greater than 0 (positive correlation)
To further examine the relationship between immune cells and LDL-C-associated intestinal microbiota, the study analyzed the connection between 22 immune cells and their dominant microbiota in the two groups. In H-LDL-C group, g__Fusobacterium.s__Fusobacterium_necrophorum was significantly positively correlated with Tregs; g__Oscillibacter.s__uncultured_bacterium , f__Shewanellaceae.g__Shewanella , o__Altero-monadales.f__Shewanellaceae , c__Gamma-proteobacteria.o__Alteromonadales , f__Coriobacteriaceae.g__Paraeggerthella , g__Paraeggerthella.s__Paraeggerthella_hongkongensis were significantly positively correlated with resting NK cells. Among these g__Paraeggerthella.s__Paraeggerthella_hongkongensis was also significantly positively correlated with Tfh and significantly negatively correlated with plasma cells(Fig. 3 B, D ). In the L-LDL-C group, g__Anaerostipes.s__Anaerostipes_caccae was significantly positively correlated with neutrophils, o__Actinomycetales.f__Corynebacteriaceae and f__Corynebacteriaceae.g__Corynebacterium were significantly negatively correlated with plasma cells; f__Veillonellaceae.g__Veillonella was significantly negatively correlated with monocytes (Fig. 3 C, D ). In summary, there were significant differences in the proportion of tumor-infiltrating immune cells between CRC patients in two groups. Additionally, several dominant gut microbiota in H-LDL-C group showed significant correlations with immune cells, suggesting that LDL-C-associated gut microbiota may influence CRC progression by regulating immune cell infiltration.
The connection between LDL-C-associated gut microbiota and immune-related genes
The immune system is pivotal in cancer progression. To examine the correlation between LDL-C-related intestinal microbiota and immune function, the study conducted a connection analysis between LDL-C related intestinal microbiota and prevalent immune- associated genes. In H-LDL-C group, the dominant gut microbiota o__Alteromonadales.f__Shewanellaceae , g__Oscillibacter.s__uncultured_bacterium , f__Shewanellaceae.g__Shewanella and c__Gammaproteobacteria.o__Alteromonadales were significantly positively correlated with multiple immune checkpoints (KIR3DL1, LAIR1, CD28, and CD80, etc.) (Fig. 4 A), chemokines (CCL7, CXCL3, and CCL3, etc.) (Fig. 4 B), immune activation genes (CD80 and CD28) (Supplementary Fig. 2), immunosuppressive genes (HAVCR2) (Supplementary Fig. 3) and chemokine receptors (XCR1) (Supplementary Fig. 4). In L-LDL-C group, the dominant gut microbiota g__Butyricimonas.s__uncultured_bacterium and f__Acidamin-ococcaceae.g__Acidaminococcus demonstrated a significant positive correlation with multiple immune checkpoints (PDCD1LG2, TNFSF14, and HAVCR2, etc.) (Fig. 4 C), chemokines (CXCL9, CCL8, CCL7, and CCL5, etc.) (Fig. 4 D), immune activating genes (TNSF14, TNFSF13B, KLRK1, and CD28, etc.) (Supplementary Fig. 5), immunosuppressive genes (PDCD1LG2 and HAVCR2, etc.) (Supplementary Fig. 6), and chemokine receptors (XCR1, CCR5, and CCR1) (Supplementary Fig. 7). These results suggest that LDL-C-associated differential gut microbiota may serve a vital function in the regulation of immune-related gene expression and the CRC progression.

Correlation between LDL-C related differential gut microbiota and immune related genes. A Heat map of the correlation between dominant gut microbiota and immune checkpoints in the H-LDL-C group. B Heat map of the correlation between dominant gut microbiota and chemokines in the H-LDL-C group. C Heat map of the correlation between dominant gut microbiota and immune checkpoints in the L-LDL-C group. D Heat map of the correlation between dominant gut microbiota and chemokines in the L-LDL-C group. The horizontal axis represents genes and the vertical axis represents gut microbiota. In the figure, red represents positive correlation, blue represents negative correlation, color depth represents the magnitude of Pearson correlation coefficient, and color from light to dark represents the value of Pearson correlation coefficient from small to large. The “*” in the figure represents the size of the P -value: no * represents P -value ≥ 0.05, * represents 0.01 ≤ P < 0.05, * * represents 0.001 ≤ P < 0.01, and * * * represents P < 0.001
Analysis of differential pathways and their connection with gut microbiota according to LDL-C levels
To further investigate the relationship between regulatory pathways associated with LDL-C and LDL-C related gut microbiota, GO and KEGG were conducted. RNA-seq data obtained from tumor specimens of 8 patients, who also underwent 16S rRNA sequencing of intestinal microbiota were converted into scoring matrices using the ssGSEA method. Figures 5 A and B show that the analysis of GO and KEGG pathway score matrices for two groups. 139 GO pathways were significantly upregulated in H-LDL-C group [GOBP_KILLING_OF_CELLS_OF_ANOTHER_ORGANISM (logFC = 0.041, P < 0.001), GOMF_EFFLUX_TRANSMEMBRANE_TRANSPORTER_ACTIVITY (logFC = -0.044, P = 0.009) and GOCC_CILIARY_TIP (logFC = -0.031, P = 0.006), etc.] as well as 2 KEGG pathways were significantly upregulated [KEGG_ALDOSTERONE_REGULATED_SODIUM_REAB-SORPTION (logFC = 0.036, P = 0.014) and KEGG_PANTOTHENATE_AND_COA_BIOSYNTHESIS (logFC = 0.040, P = 0.029) (Supplementary Tables 5 and 6 show KEGG and GO list, respectively). The findings indicate that CRC related to LDL-C metabolism exhibit distinct biological functions.

Identification of LDL-C related differential pathways and correlation between differential pathways and LDL-C related differential gut microbiota. A GO volcano plot of LDL-C related differential expression. B KEGG volcano map of LDL-C related differential expression. The horizontal coordinate represents log2 (fold change), and the further the point is from the center, the greater the differential fold; The vertical coordinate represents -log10 ( P -value), and the closer to the top point, the more significant the difference in expression. Each point represents the detected differentially expressed genes, with red indicating upregulated genes, blue indicating downregulated genes, and gray indicating no differentially expressed genes. C Correlation diagram between LDL-C related differential BP, MF pathway and differential gut microbiota. The horizontal coordinate represents microbiota, and the vertical coordinate represents GO labels. In the figure, red represents positive correlation, blue represents negative correlation, color depth represents the magnitude of Spearman correlation coefficient, and color from light to dark represents Spearman correlation coefficient value from small to large. In the figure "×" symbol represents the P -value: "×" represents P value ≥ 0.05, without "×" represents P < 0.05
To further investigate the relationship between LDL-C related genomic functions and differential gut microbiota, the study analyzed the correlation between the colony counts of 24 LDL-C-related microbiota from 8 patients and LDL-C-related BP, MF, and KEGG pathway scoring matrices. Significant correlations were observed between some differential microbiota and specific BP and MF pathways. For instance, in the H-LDL-C group, the upregulated pathway GOMF_CARBOHYDRATE_TRANSMEMBRANE_TRANSPORTER_ACTIVITY exhibited a strong positive correlation with o__Sphingobacteriales.f__Chitinophagaceae (r = 0.9, P < 0.05) and f__Coriobacteriaceae.g__Paraeggerthella (r = 0.86, P < 0.05), while GOMF_SUGAR_TRANSMEM-BRANE_TRANSPORTER_ACTIVITY showed a significant positive correlation with these two microbiota (r = 0.81, P < 0.05; r = 0.71, P < 0.05). In the L-LDL-C group, the upregulated pathway GOBP_NEGATIVE_REGULATION_OF_ENDOPLASMIC_RETICULUM_UNFOLDED_PROTEIN_RESPONSE and g__Anaerostipes.s__Anaerostipes_caccae exhibited a pronounced inverse correlation (r = 0.76, P < 0.05) (Fig. 5 C; Supplementary Tables 7 and 8). However, upregulated KEGG pathways in both groups of samples did not show significant correlations with these differential gut microbiota (Supplementary Table 9). These results suggest that LDL-C and its associated differential gut microbiota may influence CRC progression through various potential biological interactions.
Construction of biological predictive models for LDL-C status through differential intestinal microbiota
To further identify intestinal microbiota linked to LDL-C and evaluate their prognostic ability, prediction models using MLP and XGB were constructed based on 24 LDL-C related differential gut microbiota identified through LEfSe analysis.
In the MLP-based LDL-C prediction model, the training cohort confusion matrix (Fig. 6 A) indicated that the counts of true negative (TN) and true positive (TP) samples were significantly exceeding those of false negative (FN) and false positive (FP) samples. In the validation cohort (Fig. 6 B), the number of TN predictions was similar to FN predictions, while TP predictions were notably higher than FP predictions. The ROC curve analysis revealed a 0.940 AUC value for the training cohort and 0.750 for the validation cohort (Fig. 6 C).

The effectiveness evaluation of MLP and XGB prediction models. A The confusion matrix of MLP in the training set. B The confusion matrix of the MLP model in the validation set. D The confusion matrix of the XGB model in the training set. E The confusion matrix of the XGB model in the validation set. The Y-axis represents the predicted results of the model, the X-axis represents the true situation, 1 represents correct prediction, 0 represents incorrect prediction, and the value in the box represents the number of samples. C ROC curves of MLP model training and validation sets. F ROC curves of XGB prediction model training and validation sets. The horizontal axis represents the false positive rate predicted by the model, the vertical axis represents the true positive rate predicted by the model, and the area under the curve represents the AUC value. The higher the AUC value, the higher the diagnostic performance of the model
In the XGB-based LDL-C prediction model, the training cohort of confusion matrix (Fig. 6 D) showed a significantly higher number of TN and TP samples compared to FN and FP samples. The counts of TN and TP predictions in the validation cohort were greater than those of FN and FP predictions (Fig. 6 E). The XGB model’s ROC curve analysis resulted in 0.978 AUC value for the training cohort and 0.601 for the validation cohort (Fig. 6 F).
The findings imply that both models demonstrate varying levels of accuracy in predicting LDL-C status, with XGB showing superior performance in the training cohort, while MLP model demonstrated better validation cohort performance.
This study examined differences in gut microbiota between CRC patients in H-LDL-C and L-LDL-C groups. The study employed 16S rRNA sequencing to assess the composition and abundance of intestinal microbiota associated with LDL-C in CRC patients. It identified key microbiota essential for distinguishing LDL-C metabolic disorders and used these findings to examine microbial factors related to LDL-C metabolism disorders, interactions among microbial communities, and the causes of microbial variation in CRC patients. Meanwhile, the study investigated the TME and biological functions, and used immune characteristic analysis to investigate the association between particular intestinal microbiota and CRC. Further analyses were performed to evaluate the biological effects of varying microbiota and LDL-C metabolism on CRC progression.
Although HDL-C and TG levels showed no significant differences between two groups, a higher proportion of patients in the H-LDL-C group had abnormal serum total cholesterol levels. The use of the LDL-C regulatory drug Evolocumab, as demonstrated by Koskinas KC et al. to lower LDL-C concentrations in acute coronary syndrome patients, resulted in significant reductions in total cholesterol levels. This suggests that abnormal LDL-C metabolism may play a crucial role in increasing serum total cholesterol levels in CRC patients [ 24 ]. LDL-C can promote CRC cell proliferation by regulating lipid metabolism within CRC cells. Additionally, the connection between high cholesterol levels and increased CRC risk further supports the notion that abnormal LDL-C metabolism may be crucial in CRC development [ 25 , 26 ]. Dynamic monitoring of LDL-C level changes in suspected and high-risk CRC patients could be valuable for tracking disease progression. When examining abnormal LDL-C metabolism, the comparison of microbial diversity between CRC patients in two groups revealed no significant differences in fecal microbiota diversity within or between groups. Similarly, the research conducted by Fu J and colleagues did not find a correlation between gut microbiota and LDL-C levels [ 27 ]. This indicates that research on LDL-C-related microbial diversity may need larger sample sizes. Despite the lack of significant diversity results, in this study, PLS-DA analysis revealed notable intergroup distinguishability in gut microbiota. Although further studies are needed to resolve this contradiction, these results indicate that alterations in gut microbiota are linked to LDL-C metabolism.
Whilst, CRC is linked to alterations in diverse intestinal microbiota, including Fusobacterium nucleatum , Peptostreptococcus stomatis , and other microbiota [ 13 ]. Therefore, further analysis was conducted on gut microbiota with significant differences in abundance between CRC patients in two groups.
LEfSe analysis showed that Shewanella had a higher abundance in the H-LDL-C group of CRC patients [ 28 ]. Shewanella ’s unique fatty acid system can produce various fatty acids with a low melting point, including monounsaturated fatty acids (MUFA) and branched-chain fatty acids (BCFA) [ 29 ]. When MUFA was used instead of saturated fatty acids, an increase in MUFA intake led to a synchronous decrease in plasma cholesterol concentration due to lower LDL-C levels [ 30 ]. The findings suggest increased levels of Shewanella could mark the onset of gut microbiota self-regulation against abnormal LDL-C levels. Shewanella may act as an antagonistic microbiota and a marker of abnormal LDL-C metabolism in CRC patients, potentially serving as a key indicator for managing LDL-C metabolism in these patients. Lactobacillus delbrueckii , significantly enriched in CRC patients of L-LDL-C group, can reverse elevated levels of various lipids, including LDL-C, caused by Staphylococcus aureus and Escherichia coli . Its ability to regulate host lipids has been confirmed by da Costa WKA et al. [ 31 , 32 ]. Lactobacillus delbrueckii may regulate LDL-C levels by increasing free fatty acid (FFA) levels, which mediate the redistribution of lipid regulatory pools within liver cells, ultimately leading to lower LDL-C levels [ 33 , 34 ]. This may explain why LDL-C levels did not increase abnormally in CRC patients enriched with Lactobacillus delbrueckii, suggesting its potential use as a live biotherapeutic agent for managing LDL-C metabolic disorders. Additionally, Veillonella , another differential microbiota, showed higher abundance in the L-LDL-C group and was significantly correlated with seven different microbiota. Veillonella can colonize the intestine under inflammatory conditions and is associated with CRC adenocarcinoma and chemotherapy resistance. It is highly enriched in CRC patients’ proximal colon [ 35 , 36 , 37 , 38 ]. Among the Veillonella -associated microbiota, Coprobacillus , the dominant genus in the H-LDL-C group, showed a significant increase in abundance in high-fat diet-fed mice and was positively correlated with serum LDL-C levels, while exhibiting low abundance in CRC patients [ 39 , 40 ]. Therefore, it is speculated that Veillonella negatively regulates Coprobacillus abundance under CRC conditions, thereby affecting LDL-C metabolism. In a high LDL-C environment, Coprobacillus may affect CRC development through decreasing Veillonella abundance. Although these speculations need further confirmation through wet experiments, the results indicate that interactions among microbiota could play a pivotal role in changes in LDL-C levels and disease progression in CRC patients. Investigating strategies to supplement antagonistic microbiota could provide therapeutic benefits for CRC patients.
Although the findings suggest an effect of microbiota on LDL-C and CRC, the mechanisms by which these LDL-C-associated microbiota influence CRC progression remain unclear. The close relationship between intestinal mucosal immunity and gut microbiota has sparked interest in this study. First, microbiota can accelerate the progression of intestinal diseases through affecting immune environment. For example, affected by a high-fat diet, the gut microbiota can promote KRAS mutation driven intestinal carcinogenesis by influencing Major histocompatibility complex II (MHC II) antigen presentation, thereby mediating immune escape [ 41 ]. Conversely, immune environment can also directly affect gut microbiota changes. For instance, a defect in surface receptor TLR5 of flagellin can cause unstable changes in gut microbiota and induce chronic intestinal inflammation [ 42 ].
Therefore, immune infiltration analysis was conducted on the two patient groups, revealing that Tfh and Tregs had a proportional advantage in the H-LDL-C group. This characteristic could provide an important foundation for subgroup segmentation in CRC patients undergoing immunotherapy. Research has shown that the oxidized product of LDL-C, oxLDL, can regulate Tregs apoptosis and promote the generation of Tfh by modulating Tregs receptor levels [ 43 , 44 ]. Integrating the results of this research, it can be inferred that a high LDL-C environment is a critical factor affecting the characteristic proportions of Treg and Tfh in the H-LDL-C group. Targeted regulation of LDL-C, in conjunction with CRC immunotherapy, may improve treatment efficacy for patients. Building on the inferred relationship between LDL-C and TME immune cells, the study examined the association between microbiota and immune cell characteristics. It found that Fusobacterium necrophorum , significantly positively correlated with Tregs in H-LDL-C group, was more abundant in CRC tissue. Moreover, the Fusobacterium genus can suppress T cell proliferation and trigger T cell apoptosis, thereby impairing the ability to eliminate and transform cancer cells, similar to Tregs, which inhibit anti-tumor immune function [ 45 ]. Based on the correlation between microbiota abundance and Treg infiltration, along with the consistency between microbiota and Treg function, it can be speculated that in a high LDL-C environment, Fusobacterium necrophorum could recruit Tregs through its metabolites to inhibit the anti-tumor immune response, while creating a favorable environment for its own growth via TME immune changes. Therefore, targeted supplementation of Fusobacterium necrophorum for patients with elevated LDL-C levels may slow tumor progression, aiding in achieving a complete comprehensive treatment process.
Subsequently, immune-related gene association analysis was conducted to further investigate the link between LDL-C-associated gut microbiota and immune gene alterations in CRC patients. Among various immune gene associated microbiota, g_Oscillibacter showed high abundance in healthy individuals compared to CRC patients, while g_Butyricimonas exhibited enrichment in CRC tissues [ 46 , 47 ]. Both g_Butyricimona and g_Oscillibacter showed a co-directional reduction consistent with LDL-C in a study using lactoferrin to regulate metabolic disorders in obese mice [ 48 ]. Meanwhile, g_Oscillibacter can promote white adipose tissue inflammatory response by stimulating macrophages, and intestinal inflammation significantly contributes to cancer transformation and tumor progression [ 49 ]. Among multiple immune checkpoints positively correlated with LDL-C associated microbiota, KIR is a key factor in regulating NK cell activity [ 50 ]. The immune checkpoint KIR3DL1, a gene in the KIR family, provides significant protection against metastasis and peripheral nerve invasion in CRC adenocarcinoma patients by accumulating with other KIR activation genes in the same family [ 51 ]. PD-L1 is a promising candidate for CRC immunotherapy. Both PD-L2 and PD-L1 are critical signals in the T cell proliferation activation co-stimulatory molecule family B7: CD28. The immune checkpoint PDCD1LG2, a PD-L2 coding gene, is primarily expressed in monocytes and macrophages associated with CRC tumors. It may inhibit the development of tertiary lymphoid structures formed by the inflammatory aggregation of immune cells during CRC progression [ 52 , 53 , 54 ]. In the aforementioned analysis, g_Oscillibacter exhibited a significant positive connection with KIR3DL1, while g_Butyricimonas was significantly positively correlated with PDCD1LG2. Combining previous research findings, in high LDL-C environments, g_Oscillibacter and KIR3DL1 play an antagonistic role in regulating cancer progression, jointly affecting CRC. g _Butyricimonas was positively correlated with the immune processes that inhibit CRC development. These findings further underscore the crucial role of LDL-C-related gut microbiota in CRC development. Additionally, these inferences provide insights into tumor immunotherapy and its efficacy evaluation, guided by g_Oscillibacter and g_Butyricimonas .
Subsequently, the biological role of LDL-C metabolic disorders and related gut microbiota in CRC disease was further explored through gene enrichment analysis. The KEGG pathway for pantothenate and CoA biosynthesis was elevated in H-LDL-C group. Pantothenic acid (also known as vitamin B5), a component of coenzyme A, is key in intracellular lipid metabolism [ 55 ]. The pantothenic acid and coenzyme A pathways can mediate T cell metabolic reprogramming via oxidative phosphorylation, enhancing anti-tumor activity [ 56 ]. This pathway is strongly related to CRC [ 57 ]. Additionally, the GO Biological Process pathway involved in the negative regulation of the unfolded protein response in the endoplasmic reticulum was upregulated in the L-LDL-C group. When the homeostasis of endoplasmic reticulum is disturbed by inflammation, hypoxia, or other stimuli, the unfolded protein response is initiated to restore balance. Failure to restore homeostasis results in cell apoptosis [ 58 ]. This adaptive response enhances tumor cell adaptability to hypoxic stress, leading to malignant progression [ 59 ]. The key transcription factor XBP1 in this reaction can be activated in tumor associated macrophages, promoting CRC growth and metastasis [ 60 ]. LDL-C activates the IRE1 and PERK signaling pathways in endoplasmic reticulum’s unfolded protein response [ 61 ]. In this study, the dominant microbiota g__Anaerostipes.s__Anaerostipes_caccae in L-LDL-C group was significantly negatively correlated with the GOBP_NEGATIVE_REGULATION_OF_ENDOPLASMIC_RETICULUM_UNFOLDED_PROTEIN_RESPONSE pathway. Anaerostipes can inhibit tumor growth by enhancing CD8 T cells infiltration into CRC tumor tissue [ 62 ]. This indicates that LDL-C and Anaerostipes have a dynamic equilibrium, directly or indirectly interfering with endoplasmic reticulum unfolded protein response, thus influencing CRC progression. However, LDL-C and Anaerostipes only showed a negative correlation after synbiotic supplements in lactating pigs [ 63 ]. Further research is needed to confirm this conclusion, suggesting that the combination of LDL-C and Anaerostipes may be a potential CRC screening biomarker.
In recent years, machine learning such as the multilayer perceptron (MLP) model and XGBoost (XGB) model have been applied to predict CRC clinical conditions and biochemical indexes [ 64 , 65 ]. Although LDL-C status can be obtained through serology examination, research on gut microbiota has deepened, making it increasingly useful for the prevention and diagnosis of CRC, including clinical status recognition [ 66 , 67 ]. Therefore, MLP and XGB models, combined with differential gut microbiota, were used to predict the LDL-C status of CRC patients. Overall, the confusion matrices of both models indicate some false alarm rates in their predictions. However, the AUC of the ROC curve for MLP-based LDL-C prediction model was greater than 0.7 in both training and validation cohorts. For XGB-based model, it has also demonstrated good predictive performance. The findings suggest that both models possess certain predictive precision, with the MLP-based LDL-C prediction model showing better performance. This model has practical clinical utility to detect LDL-C metabolism in CRC patients through gut microbiota. The construction of these models provides more clinical significance for CRC patients’ gut microbiota, offering biological indicators for clinical evaluation and biological treatment of LDL-C-guided CRC.
This research has some limitations, particularly the lack of a healthy population control, which hinders the comparison of LDL-C metabolic disorders and microbial status between CRC patients and healthy individuals. Future studies will include healthy populations to further investigate the relationship between characteristic microbiota in CRC patients and LDL-C metabolic disorders. However, this research aims to identify microbial factors contributing to abnormal LDL-C metabolism in CRC patients and understand the pathogenesis of these differential microbial communities. Comparing two groups of CRC patients is more effective for identifying microbiota related to LDL-C metabolic disorders within the same cancer background. Interestingly, some characteristic bacterial communities, including Veillonella , can colonize other natural human lumens connected to the gastrointestinal tract [ 68 ]. Research on the changes and connections between these privileged sites and intestinal microbiota in CRC progression will provide more accurate and personalized biological basis for diagnosing and treating CRC.
Secondly, although the study has identified the characteristic intestinal microbiota potential in improving LDL-C metabolic disorders and aiding in diagnosing and treating CRC through clinical sample data analysis and previous studies, it is essential to validate these results and inferences through wet experiments in experimental organisms. Further exploration of the mechanisms by which gut microbiota mediate LDL-C metabolic disorders and CRC progression is also necessary. Additionally, the data for this study are sourced from real clinical patients, providing irreplaceable biochemical and gut microbiota results for CRC patients. This approach can significantly reduce errors among subjects and disease characteristics, offering research guidance based on microbiota abundance to explore the connection between microbiota and metabolic disorders of LDL-C in CRC patients.
This study focused on microbial factors related to LDL-C metabolic disorders and microbial pathogenicity within the upper limit of clinical LDL-C normal values. Future research will investigate how gut microbiota differ across various LDL-C levels and their relationship with CRC pathogenicity.
The metabolic status of LDL-C in CRC patients is regulated by gut microbiota. When LDL-C levels are abnormally elevated, gut microbiota can influence immune cell function and immune gene expression within the host TME. This, in turn, affects cancer-related biological pathways and promotes CRC progression. LDL-C and its associated gut microbiota could serve as non-invasive biomarkers for CRC clinical evaluation and treatment.
Data availability
The original contributions presented in the study are included in the article material, further inquiries can be directed to the corresponding authors.

Abbreviations
16S ribosomal RNA
Low-density lipoprotein cholesterol
Colorectal cancer
Oxidized low-density lipoprotein
Squalene epoxidase
Single Sample Gene Set Enrichment Analysis
Monounsaturated fatty acids
Branched chain fatty acids
Free fatty acids
Multilayer perceptron
Partial least squares discriminant analysis
Receiver operating curve
Area under curve
Linear discriminant analysis
LDA Effect Size
Tumor microenvironment
Follicular helper T cells
Regulatory T cells
Gene Ontology
Kyoto encyclopedia of Genes and Genomes
Major histocompatibility complex II
Biological processes
Cellular components
Molecular functions
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209–49.
Article PubMed Google Scholar
Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70(3):145–64.
Després JP, Lemieux I. Abdominal obesity and metabolic syndrome. Nature. 2006;444(7121):881–7.
Bankoski A, Harris TB, McClain JJ, Brychta RJ, Caserotti P, Chen KY, Berrigan D, Troiano RP, Koster A. Sedentary activity associated with metabolic syndrome independent of physical activity. Diabetes Care. 2011;34(2):497–503.
Article PubMed PubMed Central Google Scholar
Chen H, Zheng X, Zong X, Li Z, Li N, Hur J, Fritz CD, Chapman W Jr, Nickel KB, Tipping A, Colditz GA, Giovannucci EL, Olsen MA, et al. Metabolic syndrome, metabolic comorbid conditions and risk of early-onset colorectal cancer. Gut. 2021;70(6):1147–54.
Article CAS PubMed Google Scholar
Silvente-Poirot S, Poirot M. Cholesterol epoxide hydrolase and cancer. Curr Opin Pharmacol. 2012;12(6):696–703.
Wang C, Li P, Xuan J, Zhu C, Liu J, Shan L, Du Q, Ren Y, Ye J. Cholesterol Enhances Colorectal Cancer Progression via ROS Elevation and MAPK Signaling Pathway Activation. Cellular physiology and biochemistry : international journal of experimental cellular physiology, biochemistry, and pharmacology. 2017;42(2):729–42.
Jiang J, Yan M, Mehta JL, Hu C. Angiogenesis is a link between atherosclerosis and tumorigenesis: role of LOX-1. Cardiovasc Drugs Ther. 2011;25(5):461–8.
Kwon MJ, Lee JY, Kim EJ, Ko EJ, Ryu CS, Cho HJ, Jun HH, Kim JW, Kim NK. Genetic variants of MUC4 are associated with susceptibility to and mortality of colorectal cancer and exhibit synergistic effects with LDL-C levels. PLoS ONE. 2023;18(6): e0287768.
Article CAS PubMed PubMed Central Google Scholar
Ghahremanfard F, Mirmohammadkhani M, Shahnazari B, Gholami G, Mehdizadeh J. The Valuable Role of Measuring Serum Lipid Profile in Cancer Progression. Oman Med J. 2015;30(5):353–7.
Schoen RE, Pinsky PF, Weissfeld JL, Yokochi LA, Church T, Laiyemo AO, Bresalier R, Andriole GL, Buys SS, Crawford ED, Fouad MN, Isaacs C, Johnson CC, et al. Colorectal-cancer incidence and mortality with screening flexible sigmoidoscopy. N Engl J Med. 2012;366(25):2345–57.
Löwenmark T, Löfgren-Burström A, Zingmark C, Eklöf V, Dahlberg M, Wai SN, Larsson P, Ljuslinder I, Edin S, Palmqvist R. Parvimonas micra as a putative non-invasive faecal biomarker for colorectal cancer. Sci Rep. 2020;10(1):15250.
Yu J, Feng Q, Wong SH, Zhang D, Liang QY, Qin Y, Tang L, Zhao H, Stenvang J, Li Y, Wang X, Xu X, Chen N, et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut. 2017;66(1):70–8.
Kang X, Ng SK, Liu C, Lin Y, Zhou Y, Kwong TNY, Ni Y, Lam TYT, Wu WKK, Wei H, Sung JJY, Yu J, Wong SH. Altered gut microbiota of obesity subjects promotes colorectal carcinogenesis in mice. EBioMedicine. 2023;93: 104670.
Tsoi H, Chu ESH, Zhang X, Sheng J, Nakatsu G, Ng SC, Chan AWH, Chan FKL, Sung JJY, Yu J. Peptostreptococcus anaerobius Induces Intracellular Cholesterol Biosynthesis in Colon Cells to Induce Proliferation and Causes Dysplasia in Mice. Gastroenterology. 2017;152(6):1419-1433.e1415.
Li C, Wang Y, Liu D, Wong CC, Coker OO, Zhang X, Liu C, Zhou Y, Liu Y, Kang W, To KF, Sung JJ, Yu J. Squalene epoxidase drives cancer cell proliferation and promotes gut dysbiosis to accelerate colorectal carcinogenesis. Gut. 2022;71(11):2253–65.
Wu Y, Zhang Q, Ren Y, Ruan Z. Effect of probiotic Lactobacillus on lipid profile: A systematic review and meta-analysis of randomized, controlled trials. PLoS ONE. 2017;12(6): e0178868.
Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–7.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–50.
Albaradei S, Thafar M, Alsaedi A, Van Neste C, Gojobori T, Essack M, Gao X. Machine learning and deep learning methods that use omics data for metastasis prediction. Comput Struct Biotechnol J. 2021;19:5008–18.
Ali H, Ahmed A, Olivos C, Khamis K, Liu J. Mitigating urinary incontinence condition using machine learning. BMC Med Inform Decis Mak. 2022;22(1):243.
Wang Z, Xu C, Liu W, Zhang M, Zou J, Shao M, Feng X, Yang Q, Li W, Shi X, Zang G, Yin C. A clinical prediction model for predicting the risk of liver metastasis from renal cell carcinoma based on machine learning. Front Endocrinol. 2022;13:1083569.
Article Google Scholar
Li K, Yao S, Zhang Z, Cao B, Wilson CM, Kalos D, Kuan PF, Zhu R, Wang X. Efficient gradient boosting for prognostic biomarker discovery. Bioinformatics (Oxford, England). 2022;38(6):1631–8.
CAS PubMed Google Scholar
Koskinas KC, Windecker S, Pedrazzini G, Mueller C, Cook S, Matter CM, Muller O, Häner J, Gencer B, Crljenica C, Amini P, Deckarm O, Iglesias JF, et al. Evolocumab for Early Reduction of LDL Cholesterol Levels in Patients With Acute Coronary Syndromes (EVOPACS). J Am Coll Cardiol. 2019;74(20):2452–62.
Mayengbam SS, Singh A, Yaduvanshi H, Bhati FK, Deshmukh B, Athavale D, Ramteke PL, Bhat MK. Cholesterol reprograms glucose and lipid metabolism to promote proliferation in colon cancer cells. Cancer & metabolism. 2023;11(1):15.
Yao X, Tian Z. Dyslipidemia and colorectal cancer risk: a meta-analysis of prospective studies. Cancer causes & control : CCC. 2015;26(2):257–68.
Fu J, Bonder MJ, Cenit MC, Tigchelaar EF, Maatman A, Dekens JA, Brandsma E, Marczynska J, Imhann F, Weersma RK, Franke L, Poon TW, Xavier RJ, et al. The Gut Microbiome Contributes to a Substantial Proportion of the Variation in Blood Lipids. Circ Res. 2015;117(9):817–24.
Lemaire ON, Méjean V, Iobbi-Nivol C. The Shewanella genus: ubiquitous organisms sustaining and preserving aquatic ecosystems. FEMS Microbiol Rev. 2020;44(2):155–70.
Wang F, Xiao X, Ou HY, Gai Y, Wang F. Role and regulation of fatty acid biosynthesis in the response of Shewanella piezotolerans WP3 to different temperatures and pressures. J Bacteriol. 2009;191(8):2574–84.
Gill JM, Brown JC, Caslake MJ, Wright DM, Cooney J, Bedford D, Hughes DA, Stanley JC, Packard CJ. Effects of dietary monounsaturated fatty acids on lipoprotein concentrations, compositions, and subfraction distributions and on VLDL apolipoprotein B kinetics: dose-dependent effects on LDL. Am J Clin Nutr. 2003;78(1):47–56.
Evivie SE, Abdelazez A, Li B, Bian X, Li W, Du J, Huo G, Liu F. In vitro Organic Acid Production and In Vivo Food Pathogen Suppression by Probiotic S. thermophilus and L. bulgaricus. Front Microbiol. 2019;10:782.
da Costa WKA, Brandão LR, Martino ME, Garcia EF, Alves AF, de Souza EL, de Souza AJ, Saarela M, Leulier F, Vidal H, Magnani M. Qualification of tropical fruit-derived Lactobacillus plantarum strains as potential probiotics acting on blood glucose and total cholesterol levels in Wistar rats. Food research international (Ottawa, Ont). 2019;124:109–17.
Hou G, Yin J, Wei L, Li R, Peng W, Yuan Y, Huang X, Yin Y. Lactobacillus delbrueckii might lower serum triglyceride levels via colonic microbiota modulation and SCFA-mediated fat metabolism in parenteral tissues of growing-finishing pigs. Frontiers in veterinary science. 2022;9:982349.
Daumerie CM, Woollett LA, Dietschy JM. Fatty acids regulate hepatic low density lipoprotein receptor activity through redistribution of intracellular cholesterol pools. Proc Natl Acad Sci USA. 1992;89(22):10797–801.
Rojas-Tapias DF, Brown EM, Temple ER, Onyekaba MA, Mohamed AMT, Duncan K, Schirmer M, Walker RL, Mayassi T, Pierce KA, Ávila-Pacheco J, Clish CB, Vlamakis H, et al. Inflammation-associated nitrate facilitates ectopic colonization of oral bacterium Veillonella parvula in the intestine. Nat Microbiol. 2022;7(10):1673–85.
Kasai C, Sugimoto K, Moritani I, Tanaka J, Oya Y, Inoue H, Tameda M, Shiraki K, Ito M, Takei Y, Takase K. Comparison of human gut microbiota in control subjects and patients with colorectal carcinoma in adenoma: Terminal restriction fragment length polymorphism and next-generation sequencing analyses. Oncol Rep. 2016;35(1):325–33.
Deng X, Li Z, Li G, Li B, Jin X, Lyu G. Comparison of Microbiota in Patients Treated by Surgery or Chemotherapy by 16S rRNA Sequencing Reveals Potential Biomarkers for Colorectal Cancer Therapy. Front Microbiol. 2018;9:1607.
Sheng Q, Du H, Cheng X, Cheng X, Tang Y, Pan L, Wang Q, Lin J. Characteristics of fecal gut microbiota in patients with colorectal cancer at different stages and different sites. Oncol Lett. 2019;18(5):4834–44.
CAS PubMed PubMed Central Google Scholar
Li TT, Huang ZR, Jia RB, Lv XC, Zhao C, Liu B. Spirulina platensis polysaccharides attenuate lipid and carbohydrate metabolism disorder in high-sucrose and high-fat diet-fed rats in association with intestinal microbiota. Food research international (Ottawa, Ont). 2021;147:110530.
Yang J, Li D, Yang Z, Dai W, Feng X, Liu Y, Jiang Y, Li P, Li Y, Tang B, Zhou Q, Qiu C, Zhang C, et al. Establishing high-accuracy biomarkers for colorectal cancer by comparing fecal microbiomes in patients with healthy families. Gut microbes. 2020;11(4):918–29.
Schulz MD, Atay C, Heringer J, Romrig FK, Schwitalla S, Aydin B, Ziegler PK, Varga J, Reindl W, Pommerenke C, Salinas-Riester G, Böck A, Alpert C, et al. High-fat-diet-mediated dysbiosis promotes intestinal carcinogenesis independently of obesity. Nature. 2014;514(7523):508–12.
Carvalho FA, Koren O, Goodrich JK, Johansson ME, Nalbantoglu I, Aitken JD, Su Y, Chassaing B, Walters WA, González A, Clemente JC, Cullender TC, Barnich N, et al. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe. 2012;12(2):139–52.
Gaddis DE, Padgett LE, Wu R, McSkimming C, Romines V, Taylor AM, McNamara CA, Kronenberg M, Crotty S, Thomas MJ, Sorci-Thomas MG, Hedrick CC. Apolipoprotein AI prevents regulatory to follicular helper T cell switching during atherosclerosis. Nat Commun. 2018;9(1):1095.
Li Q, Wang Y, Li H, Shen G, Hu S. Ox-LDL influences peripheral Th17/Treg balance by modulating Treg apoptosis and Th17 proliferation in atherosclerotic cerebral infarction. Cell Physiol Biochemistry. 2014;33(6):1849–62.
Article CAS Google Scholar
King M, Hurley H, Davidson KR, Dempsey EC, Barron MA, Chan ED, Frey A. The Link between Fusobacteria and Colon Cancer: a Fulminant Example and Review of the Evidence. Immune network. 2020;20(4):e30.
Loke MF, Chua EG, Gan HM, Thulasi K, Wanyiri JW, Thevambiga I, Goh KL, Wong WF, Vadivelu J. Metabolomics and 16S rRNA sequencing of human colorectal cancers and adjacent mucosa. PLoS ONE. 2018;13(12):e0208584.
Wu M, Wu Y, Deng B, Li J, Cao H, Qu Y, Qian X, Zhong G. Isoliquiritigenin decreases the incidence of colitis-associated colorectal cancer by modulating the intestinal microbiota. Oncotarget. 2016;7(51):85318–31.
Li L, Ma C, Hurilebagen, Yuan H, Hu R, Wang W and Weilisi. Effects of lactoferrin on intestinal flora of metabolic disorder mice. BMC Microbiol. 2022;22(1):181.
Gaudino SJ, Singh A, Huang H, Padiadpu J, Jean-Pierre M, Kempen C, Bahadur T, Shiomitsu K, Blumberg R, Shroyer KR, Beyaz S, Shulzhenko N, Morgun A, et al. Intestinal IL-22RA1 signaling regulates intrinsic and systemic lipid and glucose metabolism to alleviate obesity-associated disorders. Nat Commun. 2024;15(1):1597.
Dębska-Zielkowska J, Moszkowska G, Zieliński M, Zielińska H, Dukat-Mazurek A, Trzonkowski P and Stefańska K. KIR receptors as key regulators of NK cells activity in health and disease. Cells. 2021;10(7):1777.
Barani S, Hosseini SV, Ghaderi A. Activating and inhibitory killer cell immunoglobulin like receptors (KIR) genes are involved in an increased susceptibility to colorectal adenocarcinoma and protection against invasion and metastasis. Immunobiology. 2019;224(5):681–6.
Yang Z, Wu G, Zhang X, Gao J, Meng C, Liu Y, Wei Q, Sun L, Wei P, Bai Z, Yao H, Zhang Z. Current progress and future perspectives of neoadjuvant anti-PD-1/PD-L1 therapy for colorectal cancer. Front Immunol. 2022;13:1001444.
Lv J, Jiang Z, Yuan J, Zhuang M, Guan X, Liu H, Yin Y, Ma Y, Liu Z, Wang H, Wang X. Pan-cancer analysis identifies PD-L2 as a tumor promotor in the tumor microenvironment. Front Immunol. 2023;14:1093716.
Masugi Y, Nishihara R, Hamada T, Song M, da Silva A, Kosumi K, Gu M, Shi Y, Li W, Liu L, Nevo D, Inamura K, Cao Y, et al. Tumor PDCD1LG2 (PD-L2) Expression and the Lymphocytic Reaction to Colorectal Cancer. Cancer Immunol Res. 2017;5(11):1046–55.
Naquet P, Kerr EW, Vickers SD, Leonardi R. Regulation of coenzyme A levels by degradation: the ‘Ins and Outs.’ Prog Lipid Res. 2020;78:101028.
St Paul MSS, Han S, Israni-Winger K, Lien SC, Laister RC, Sayad A, Penny S, Amaria RN, Haydu LE, Garcia-Batres CR, Kates M, Mulder DT, Robert-Tissot C, Gold MJ, Tran CW, Elford AR, Nguyen LT, Pugh TJ, Pinto DM, Wargo JA, Ohashi PS. Coenzyme A fuels T cell anti-tumor immunity. Cell Metab. 2021;33(12):2415–27.
Yi Y, Wang J, Liang C, Ren C, Lian X, Han C, Sun W. LC-MS-based serum metabolomics analysis for the screening and monitoring of colorectal cancer. Front Oncol. 2023;13:1173424.
Bravo R, Parra V, Gatica D, Rodriguez AE, Torrealba N, Paredes F, Wang ZV, Zorzano A, Hill JA, Jaimovich E, Quest AF, Lavandero S. Endoplasmic reticulum and the unfolded protein response: dynamics and metabolic integration. Int Rev Cell Mol Biol. 2013;301:215–90.
Bi M, Naczki C, Koritzinsky M, Fels D, Blais J, Hu N, Harding H, Novoa I, Varia M, Raleigh J, Scheuner D, Kaufman RJ, Bell J, et al. ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J. 2005;24(19):3470–81.
Zhao Y, Zhang W, Huo M, Wang P, Liu X, Wang Y, Li Y, Zhou Z, Xu N, Zhu H. XBP1 regulates the protumoral function of tumor-associated macrophages in human colorectal cancer. Signal Transduct Target Ther. 2021;6(1):357.
Guevara-Olaya L, Chimal-Vega B, Castañeda-Sánchez CY, López-Cossio LY, Pulido-Capiz A, Galindo-Hernández O, Díaz-Molina R, Ruiz Esparza-Cisneros J and García-González V. LDL promotes disorders in β-cell cholesterol metabolism, implications on insulin cellular communication mediated by EVs. Metabolites. 2022;12(8):754.
Montalban-Arques A, Katkeviciute E, Busenhart P, Bircher A, Wirbel J, Zeller G, Morsy Y, Borsig L, Glaus Garzon JF, Müller A, Arnold IC, Artola-Boran M, Krauthammer M, et al. Commensal Clostridiales strains mediate effective anti-cancer immune response against solid tumors. Cell Host Microbe. 2021;29(10):1573-1588.e1577.
Ma C, Gao Q, Zhang W, Zhu Q, Tang W, Blachier F, Ding H, Kong X. Supplementing Synbiotic in Sows; Diets Modifies Beneficially Blood Parameters and Colonic Microbiota Composition and Metabolic Activity in Suckling Piglets. Frontiers in veterinary science. 2020;7:575685.
Qin L, Liang Z, Xie J, Ye G, Guan P, Huang Y, Li X. Development and validation of machine learning models for postoperative venous thromboembolism prediction in colorectal cancer inpatients: a retrospective study. Journal of gastrointestinal oncology. 2023;14(1):220–32.
Du G, Ren C, Wang J, Ma J. The Clinical Value of Blood miR-654-5p, miR-126, miR-10b, and miR-144 in the Diagnosis of Colorectal Cancer. Comput Math Methods Med. 2022;2022:8225966.
Coker OO, Liu C, Wu WKK, Wong SH, Jia W, Sung JJY, Yu J. Altered gut metabolites and microbiota interactions are implicated in colorectal carcinogenesis and can be non-invasive diagnostic biomarkers. Microbiome. 2022;10(1):35.
Fong W, Li Q, Yu J. Gut microbiota modulation: a novel strategy for prevention and treatment of colorectal cancer. Oncogene. 2020;39(26):4925–43.
Giacomini JJ, Torres-Morales J, Dewhirst FE, Borisy GG, Mark Welch JL. Site Specialization of Human Oral Veillonella Species. Microbiology spectrum. 2023;11(1):e0404222.
Download references
Acknowledgements
Natural Science Foundation of Guangxi Province (Guangxi Natural Science Foundation) (2021GXNSFAA196008); Youth Science Foundation of Guangxi Medical University (GXMUYSF202402); Youth Research Project of Guangxi Medical University Affiliated Cancer Hospital (yuanqingji2023-10hao); Middle-aged and Young Teachers’ Basic Ability Promotion Project of Guangxi (Basic Ability Promotion Project of Guangxi) (2021KY0087); China Postdoctoral Science Foundation (2023MD734155); Youth Science Foundation of Guangxi Medical University (GXMUYSF202357). Guangxi Key Laboratory of Basic and Translational Research for Colorectal Cancer. Chinese National Natural Science Foundation (82460610).
Author information
Mingjian Qin, Zigui Huang and Yongqi Huang contributed equally to this work.
Authors and Affiliations
Division of Colorectal & Anal Surgery, Department of Gastrointestinal Surgery, Guangxi Medical University Cancer Hospital, Nanning, The People’s Republic of China
Mingjian Qin, Zigui Huang, Yongqi Huang, Xiaoliang Huang, Chuanbin Chen, Yongzhi Wu, Zhen Wang, Fuhai He, Binzhe Tang, Chenyan Long, Xianwei Mo, Jungang Liu & Weizhong Tang
You can also search for this author in PubMed Google Scholar
Contributions
M. Q, Y. H, Z.H, X.H, J.L, W. T, X.M: conceived and designed the experiments; J.L, X.H, M. Q, Y.H, Z. H, C.C, Z.W, F.H, C.L, Y. W, B.T, X.M, W.T: analyzed the data; J.L, X.H, M.Q, Z.W, C. C, Y.H, Z.H, F. H, Y. W, C. L, B.T, X.M, W.T: helped with reagents/materials/analysis tools; J.L, M.Q, Y.H, Z. H, X.H, C. C, Z.W, F.H, Y.W, C. L, B.T, X.M, W.T: contributed to the writing of the manuscript. All authors reviewed the manuscript.
Corresponding authors
Correspondence to Xianwei Mo , Jungang Liu or Weizhong Tang .
Ethics declarations
Ethics approval and consent to participate.
This study was approved by the Ethics and Human Subject Committee of Guangxi Medical University Cancer Hospital.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
12944_2024_2333_moesm1_esm.pdf.
Supplementary Material 1: Supplementary Fig. 1. Box plot of KEGG functional abundance in the H-LDL-C group versus the L-LDL-C group of CRC patients. Horizontal coordinates indicate groupings, vertical coordinates indicate predicted abundance values for that pathway in each sample, boxes indicate the 25th-75th percentiles, and the center marker indicates the median; black bars are 1.5 times the interquartile range.
12944_2024_2333_MOESM2_ESM.pdf
Supplementary Material 2: Supplementary Fig. 2. Heat map of the correlation between the dominant flora of H-LDL-C group and immune activation genes.
12944_2024_2333_MOESM3_ESM.pdf
Supplementary Material 3: Supplementary Fig. 3. Heat map of the correlation between the dominant flora of H-LDL-C group and Immunosuppressive genes.
12944_2024_2333_MOESM4_ESM.pdf
Supplementary Material 4: Supplementary Fig. 4. Heat map of the correlation between the dominant flora of H-LDL-C group and chemokine receptors.
12944_2024_2333_MOESM5_ESM.pdf
Supplementary Material 5: Supplementary Fig. 5. Heat map of the correlation between the dominant flora of L-LDL-C group and immune activation genes.
12944_2024_2333_MOESM6_ESM.pdf
Supplementary Material 6: Supplementary Fig. 6. Heat map of the correlation between the dominant flora of L-LDL-C group and Immunosuppressive genes.
12944_2024_2333_MOESM7_ESM.pdf
Supplementary Material 7: Supplementary Fig. 7. Heat map of the correlation between the dominant flora of L-LDL-C group and chemokine receptors. Horizontal coordinates are genes, vertical coordinates are colonies, red in the graph represents positive correlation, blue represents negative correlation, color depth represents Pearson correlation coefficient size, color from light to dark indicates Pearson correlation coefficient value from small to large. The “*” in the graph indicates the size of P value: no * for P value ≥ 0.05, * for 0.01 ≤ P < 0.05, ** for 0.001 ≤ P < 0.01, *** for P < 0.001.
12944_2024_2333_MOESM8_ESM.docx
Supplementary Material 8: Supplementary Table 1. ADONIS test for Bray Distance of intestinal flora in CRC patients in the H-LDL-C and L-LDL-C groups.
12944_2024_2333_MOESM9_ESM.docx
Supplementary Material 9: Supplementary Table 2. ADONIS test for Jaccard Distance of intestinal flora in CRC patients in the H-LDL-C and L-LDL-C groups. Group row: between-group statistics; Residuals row: within-group statistics; Total row: between-group + within-group statistics; Df: degrees of freedom, between-group degrees of freedom are the number of groups—1, within-group degrees of freedom are the total number of samples—number of groups; Sums Of Sqs: sum of squared deviations; Mean Sqs: mean square, the ratio of sums of squared deviations to degrees of freedom, i.e. Sums Of Sqs/ Df; F.Model: F-test value, i.e. between-group mean square/within-group mean square; R2: ratio of between- and within-group sums of squared deviations to total sums of squared deviations, indicating the degree of explanation of differences between samples, with larger R2 indicating a higher degree of explanation of differences between samples; Pr (> F): statistically significant P -value obtained from the substitution test, with Pr < 0.05 as statistically different.
12944_2024_2333_MOESM10_ESM.docx
Supplementary Material 10: Supplementary Table 3. Results of LEfSe analysis. Taxonomy: information of differential species; Group: group with significant abundance of differential species; LDA: effect value of differential species; the table shows species with LDA score (log10) greater than the preset value (default is 2) and P value less than 0.05.
12944_2024_2333_MOESM11_ESM.docx
Supplementary Material 11: Supplementary Table 4. KEGG functional pathways in the intestinal microbiome of CRC patients in the H-LDL-C and L-LDL-C groups. KEGG_Pathway: KEGG pathway; Mean In H-LDL-C: the predicted abundance value of this pathway in each sample in the H-LDL-C group; Mean In L-LDL-C: the predicted abundance value of this pathway in each sample in the L-LDL-C group.
12944_2024_2333_MOESM12_ESM.docx
Supplementary Material 12: Supplementary Table 5. List of differential KEGG pathways of CRC patients stratified by LDL-C condition. FC in logFC is fold change, which indicates the ratio of H-LDL-C group to L-LDL-C expression (and takes the logarithm of its base at 2), with P -value < 0.05 as statistically significant difference.
12944_2024_2333_MOESM13_ESM.docx
Supplementary Material 13: Supplementary Table 6. List of differential GO items of CRC patients stratified by LDL-C condition. FC in logFC, i.e. fold change, denotes the ratio of H-LDL-C group to L-LDL-C expression and was taken as logarithm with a base of 2. P -value < 0.05 was taken as statistically significant difference.
12944_2024_2333_MOESM14_ESM.docx
Supplementary Material 14: Supplementary Table 7. Correclation between enrich GO items of CRC patients in H-LDL-C group and LDL-C-associated differential intestinal microbiome.
12944_2024_2333_MOESM15_ESM.docx
Supplementary Material 15: Supplementary Table 8. Correlation between enriched GO items of CRC patients in L-LDL-C group and LDL-C-associated differential intestinal microbiome.
12944_2024_2333_MOESM16_ESM.docx
Supplementary Material 16: Supplementary Table 9. Correlation between KEGG pathways of CRC patients and LDL-C-associated differential intestinal microbiome. The r.value is the Spearman correlation coefficient value, with P -value < 0.05 as statistically significant difference.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/ .
Reprints and permissions
About this article
Cite this article.
Qin, M., Huang, Z., Huang, Y. et al. Association analysis of gut microbiota with LDL-C metabolism and microbial pathogenicity in colorectal cancer patients. Lipids Health Dis 23 , 367 (2024). https://doi.org/10.1186/s12944-024-02333-4
Download citation
Received : 30 July 2024
Accepted : 17 October 2024
Published : 08 November 2024
DOI : https://doi.org/10.1186/s12944-024-02333-4
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Colorectal cancer (CRC)
- Gut microbiota
- Machine learning
- Clinical status
Lipids in Health and Disease
ISSN: 1476-511X
- General enquiries: [email protected]
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- My Account Login
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 07 November 2024
Engineered PsCas9 enables therapeutic genome editing in mouse liver with lipid nanoparticles
- Dmitrii Degtev ORCID: orcid.org/0009-0009-7131-510X 1 na1 ,
- Jack Bravo ORCID: orcid.org/0000-0003-0456-0753 2 na1 ,
- Aikaterini Emmanouilidi 1 ,
- Aleksandar Zdravković ORCID: orcid.org/0000-0002-9494-9350 1 ,
- Oi Kuan Choong ORCID: orcid.org/0000-0003-0257-4748 1 ,
- Julia Liz Touza 3 ,
- Niklas Selfjord ORCID: orcid.org/0009-0008-5628-5849 1 ,
- Isabel Weisheit 1 ,
- Margherita Francescatto 4 ,
- Pinar Akcakaya ORCID: orcid.org/0000-0001-8413-8995 1 ,
- Michelle Porritt ORCID: orcid.org/0000-0003-1700-0085 1 ,
- Marcello Maresca ORCID: orcid.org/0000-0003-0796-661X 1 ,
- David Taylor ORCID: orcid.org/0000-0002-6198-1194 2 , 5 , 6 &
- Grzegorz Sienski ORCID: orcid.org/0000-0002-2730-7710 1
Nature Communications volume 15 , Article number: 9173 ( 2024 ) Cite this article
7 Altmetric
Metrics details
- CRISPR-Cas9 genome editing
- Cryoelectron microscopy
Clinical implementation of therapeutic genome editing relies on efficient in vivo delivery and the safety of CRISPR-Cas tools. Previously, we identified PsCas9 as a Type II-B family enzyme capable of editing mouse liver genome upon adenoviral delivery without detectable off-targets and reduced chromosomal translocations. Yet, its efficacy remains insufficient with non-viral delivery, a common challenge for many Cas9 orthologues. Here, we sought to redesign PsCas9 for in vivo editing using lipid nanoparticles. We solve the PsCas9 ribonucleoprotein structure with cryo-EM and characterize it biochemically, providing a basis for its rational engineering. Screening over numerous guide RNA and protein variants lead us to develop engineered PsCas9 (ePsCas9) with up to 20-fold increased activity across various targets and preserved safety advantages. We apply the same design principles to boost the activity of FnCas9, an enzyme phylogenetically relevant to PsCas9. Remarkably, a single administration of mRNA encoding ePsCas9 and its guide formulated with lipid nanoparticles results in high levels of editing in the Pcsk9 gene in mouse liver, a clinically relevant target for hypercholesterolemia treatment. Collectively, our findings introduce ePsCas9 as a highly efficient, and precise tool for therapeutic genome editing, in addition to the engineering strategy applicable to other Cas9 orthologues.
Similar content being viewed by others
In vivo CRISPR base editing of PCSK9 durably lowers cholesterol in primates

Lung and liver editing by lipid nanoparticle delivery of a stable CRISPR–Cas9 ribonucleoprotein
In vivo adenine base editing of PCSK9 in macaques reduces LDL cholesterol levels
Introduction.
The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) systems serve as adaptive antiviral immunity mechanisms in bacteria, archaea, and large bacteriophages 1 , 2 , 3 . The CRISPR-associated (Cas) nucleases, repurposed for mammalian genome modification, can be programmed with single guide RNAs to target specific loci and introduce DNA double-stranded breaks (DSBs) 4 , 5 , 6 , 7 , 8 . These are subsequently repaired by the cellular DNA repair machinery, primarily through the non-homologous end-joining (NHEJ) pathway, leading to small insertions and deletions 5 , 6 , 8 , 9 , 10 , 11 . Alternatively, when presented with a DNA molecule homologous to the DSB locus, cells can employ homology-directed repair (HDR) to insert it into the genome 12 , 13 , 14 . Cas nucleases also form the basis for advanced genome editing approaches such as Base Editing 15 , 16 , Prime Editing 17 , and Epigenome Editing 18 , 19 , 20 . The wide applicability of these tools includes cell line engineering 5 , 6 , 8 , animal model development 21 , 22 , 23 , genetic screens 24 , 25 , 26 , cell therapies for cancer 27 , 28 , 29 , and curative treatments for genetic disorders 30 , 31 .
RNA-guided nucleases derived from CRISPR systems 32 , 33 , along with the recently identified OMEGA systems 34 , 35 , 36 , comprise a diverse toolkit for genome editing in human cells and in vivo. Streptococcus pyogenes Cas9 (SpCas9), the most studied Cas-enzyme, has proven to be a highly efficient and versatile tool for genome editing, becoming the gold standard in the field. SpCas9 was employed in clinical trials for gene therapy and in the first ex vivo genome edited cell therapy, that was recently approved by the FDA 30 , 31 , 37 . However, its promiscuous activity remains a concern for the wider application of genome editing in the clinic 38 , 39 , 40 , 41 , 42 , 43 , 44 , 45 , 46 , 47 , 48 , 49 , 50 . In response, several studies rationally engineered SpCas9 using targeted mutagenesis or applied directed evolution to eliminate its off-target activity 51 , 52 , 53 , 54 , 55 , 56 , 57 , 58 , 59 . Moreover, other natural and engineered Cas9 nucleases with high-fidelity properties from other prokaryotic species were introduced and suggested as additional tools for genome editing 60 , 61 , 62 , 63 , 64 , 65 , 66 , 67 , 68 . Our previous work characterized PsCas9 as an intrinsically high-fidelity enzyme of the Type II-B subfamily 69 . Viral delivery of PsCas9 to mouse liver in vivo resulted in high levels of editing with negligible off-target events and fewer chromosomal translocations, thus offering a safer alternative for therapeutic applications compared to SpCas9.
In this study, we thoroughly interrogate PsCas9 function through cellular, structural, and biochemical studies and engineer it for therapeutic genome editing applications. We found that the editing activity of PsCas9 is hampered in conditions of limited intracellular concentration, and it exhibits a relatively low affinity to DNA in vitro compared to SpCas9. We propose that the low editing activity of PsCas9 in cells could be rescued by improving its affinity to DNA. To this end, we solve a high-resolution structure of PsCas9 using cryo-electron microscopy (cryo-EM), thereby providing a foundation for the rational engineering of PsCas9. Optimization of the PsCas9 sgRNA scaffold results in a modest increase in its genome editing efficacy, while targeted enzyme mutagenesis leads to a remarkable improvement (up to 20-fold) across a wide set of genomic targets. These enhancements in gene editing activity in cells are correlated with more efficient DNA interaction in vitro. Importantly, the high-fidelity properties of the wild-type enzyme, including a favourable off-target profile and low translocation frequency, are preserved upon engineering. Employing a similar strategy, we modify the well-established member of Type II-B subfamily Francisella novicida Cas9 (FnCas9) 62 and increase its activity up to 15-fold. Finally, we evaluate the performance of the engineered version of PsCas9, ePsCas9, using lipid nanoparticle (LNP) delivery to mouse liver to disrupt the Pcsk9 gene, a clinically relevant approach for hypercholesterolemia treatment 70 , 71 . We demonstrate that ePsCas9 induces a high level of editing in the liver and a concurrent decrease of Pcsk9 levels in blood plasma. Collectively, this work positions ePsCas9 as a promising tool for safe and effective in vivo genome editing applications, as well as proposes an engineering strategy to enhance the activity of other Cas9 orthologs to further expand the CRISPR toolbox for medical use.
PsCas9 editing activity is limited by its intracellular concentration
We previously characterized PsCas9, a member of the Type II-B family, as a highly active and precise enzyme 69 . PsCas9 recognizes the same NGG PAM as SpCas9, and thus, can be conveniently benchmarked against it. In line with our previous work, PsCas9 induces genome editing in HEK293T cells with high efficiency (up to 80%) and comparable to SpCas9 at two tested targets (EMX1a and PCSK9) when delivered via plasmid vectors (Fig. 1a ). This delivery approach enables sustained, high expression levels of Cas9 and its sgRNA, providing excessive amounts of functional ribonucleoproteins (RNPs), and thus, high levels of editing. To assess the activity of these enzymes in a more controlled setting, we transfected cells with plasmids overexpressing the Cas9 protein only, and then introduced synthetic sgRNA in varying quantities, thereby restricting the intracellular concentration of active Cas9 RNPs. As expected, editing efficiency increased with higher doses of delivered sgRNA for both enzymes (Fig. 1b, c ). However, in stark contrast to our earlier observations, PsCas9 exhibited an order of magnitude lower efficacy than SpCas9 across all tested conditions. This finding suggests that the genome editing activity of PsCas9 is limited when the RNP abundance is constrained in the cell.

a Genome editing activity of SpCas9 and PsCas9 in HEK293T cells mediated via plasmid DNA transfection. Editing efficiency was evaluated as the percentage of reads with indels using amplicon sequencing. Data are shown as mean ± SD for n = 3 biological repeats. b , c Genome editing activity of SpCas9 and PsCsa9 in HEK293T cells mediated via plasmid DNA encoding Cas9s transfection followed by synthetic sgRNA transfection at EMX1a ( b ) and PCSK9 ( c ) sites. Editing efficiency was evaluated using amplicon sequencing. Data are shown as mean ± SD for n = 3 biological repeats. d Schematic of in vitro binding experiment of Cas9 to dsDNA “target substrate”. 38 bp substrate contains a single target site (blue) and PAM (orange). The remaining DNA (grey) was depleted of PAMs. DNA was labelled with FAM on the 3’-end of the bottom strand. e , f Sp- and PsCas9 interaction with target substrates assessed by fluorescence polarization of FAM fluorophore. The signal was fitted to a 4-parametric logistic function and binding constant Kd was extracted. Data are shown as mean ± SD for n = 3 technical replicates. g Schematic of in vitro binding experiment of Cas9 to dsDNA “no-target substrate” lacking target site. 80 bp substrate contains 17 or 19 PAMs distributed across it (orange) for EMX1a and PCSK9, respectively. DNA was labelled with FAM on the 3’-end of the bottom strand. h , i Sp- and PsCas9 interaction with no-target substrates assessed by fluorescence polarization of FAM fluorophore. Data are shown as mean ± SD for n = 3 technical replicates.
We hypothesized that the reduced editing activity of PsCas9 could be attributed to its biochemical properties. We have previously demonstrated that PsCas9 RNP cleaves DNA targets at rates similar to SpCas9 in vitro 69 . Therefore, we next examined the target DNA binding properties of PsCas9 using a fluorescence polarization assay. For this, we designed fluorescent double-stranded (ds) DNA substrates containing a single target site (EMX1a or PCSK9) and measured their interaction with Ps- or SpCas9 loaded with the respective sgRNAs (Fig. 1d ). Concentration-dependent increases in fluorescence polarization signal were observed for both Cas9 RNPs (Fig. 1e, f ). SpCas9 demonstrated strong interaction with both substrates, exhibiting an affinity of around 10 nM — a value comparable to previous reports using orthogonal binding assays 72 , 73 , 74 . In contrast, the binding affinity of PsCas9 to DNA was ~10- and 2-fold weaker for EMX1a and PCSK9 substrates, respectively. This finding motivated us to investigate the binding mode of PsCas9 in more detail.
Cas9 RNP interrogation of DNA in search of PAMs is a crucial step preceding target recognition 72 . We used the fluorescence polarization assay to evaluate PsCas9 interaction with a DNA substrate containing PAM sequences but no target 75 . This method enables the assessment of Cas9 RNP interaction with DNA through transient binding to PAMs only. We designed fluorescent dsDNA EMX1a and PCSK9 substrates, containing 17 and 19 GG dinucleotide PAMs spread across the sequence, respectively; and measured their interaction with Cas9 RNPs loaded with non-matching sgRNAs (Fig. 1g ). SpCas9 displayed a significant concentration-dependent increase in FP signal, indicating active DNA interrogation and PAM interaction (Fig. 1h, i ). In contrast, PsCas9 showed almost no increase suggesting its PAM binding is weak.
Collectively, our findings suggest that diminished PsCas9 editing activity in conditions when RNP abundance is limited can be a result of its relatively weak binding to DNA, in particular, at the interrogation step. We hypothesize that PsCas9 activity could be boosted upon strengthening its interaction with DNA. Thus, we sought to supplement our in vitro data with structural studies of PsCas9 RNP and use them for rational engineering of the enzyme for improved activity.
Cryo-EM structure of PsCas9
To understand the mechanisms of DNA recognition by PsCas9, we prepared a complex of PsCas9 with its cognate sgRNA and EMX1a dsDNA and acquired a cryo-EM dataset (Supplementary Data 1 ). During data collection, we noticed that the complex adopts a preferred orientation in ice, which we overcame by tilting the stage by −30° for the remainder of movie acquisition. After rounds of 2D and 3D classification, we obtained a cryo-EM reconstruction of PsCas9 at a global resolution of 2.9 Å (Supplementary Fig. 1 ). The quality of our map was sufficient for de novo modelling of the complete complex, including 1375 of the 1409 amino acids, the full 22-bp R-loop, and 121-nt of the sgRNA (Fig. 2a–c ). The final 10-nt of the sgRNA (positions 122 onwards) were not resolved in the map, likely due to flexibility.

a Domain architecture of PsCas9. b 2.9 Å-resolution cryo-EM reconstruction of PsCas9 in the productive state. Both target and non-target strands (TS and NTS, respectively) have been cleaved, and the HNH domain remains positioned at the scissile phosphate of the TS. c Cartoon representation of PsCas9 model. d Structural basis of NGG PAM recognition. e Comparison of PsCas9 (green cartoon) with FnCas9 (orange cartoon, PDB ID 5B2O). The crystal structure of FnCas9 represents a dead-end, non-productive state, where the HNH domain is positioned far from the cleavage position of the TS. In the productive state, HNH is shifted by up to ~60 Å to the centre of the target strand. The REC and NUC lobes are largely consistent otherwise.
Akin to previously determined type II-A, -B, and -C CRISPR effector nucleases 60 , 62 , 76 , 77 , 78 , PsCas9 has a typical bilobed architecture, with the REC1, REC2 and REC3 domains constituting the REC lobe, and the Wedge, PI, RuvC and HNH domains constituting the NUC lobe (Fig. 2a, b ). The sgRNA follows a tortuous path, interweaving between the flexibly tethered REC domains. Due to conformational heterogeneity, the REC2 domain is poorly resolved, as it has also been observed for SpCas9 58 , 79 .
Since the PAM site of the EMX1a dsDNA target is excellently resolved in our reconstruction, we could unambiguously model the PsCas9 residues responsible for PAM recognition – R1316 and R1369. These arginine residues make a bidentate interaction with the Hoogsteen faces of the two guanosine bases in the NGG motif (Fig. 2d ). While this is consistent with the mechanism of PAM readout by SpCas9 76 , 77 , the two arginine residues used by SpCas9 (R1333 and R1335) are much closer together in sequence space as in comparison to PsCas9. A similar observation was documented for another Type II-B family member, FnCas9 62 . This suggests that despite a lack of sequence homology in this region, SpCas9 and PsCas9 have evolutionarily converged on a conserved mechanism to recognise a 5’-NGG-3’ PAM motif.
Within our reconstruction, the HNH domain is docked at the target strand (TS), and the scissile phosphate of the 3 rd TS position has been cleaved, indicating that our structure corresponds to the active, productive state. A previously determined structure of FnCas9 is in a non-productive state, with both DNA strands being intact 62 (Fig. 2e ). Comparison of our productive state structure with this non-productive state homologue reveals that the HNH domain must be repositioned by up to ~ 60 Å to successfully perform catalysis. Apart from the HNH domain, the overall structural architecture of the complexes showed a high degree of similarity between FnCas9 and PsCas9 (Fig. 2e ). We observed a well-resolved density for two Mg 2+ ions in the HNH and RuvC active sites and both target and non-target strands (NTS) are cleaved (Supplementary Fig. 1f ). While structures of type II-A and -C Cas9 enzymes have been determined with both active sites in the productive state 58 , 80 , 81 , our structure represents the structure of a type II-B Cas9 enzyme in a bona fide productive state.
Type II CRISPR effector nucleases typically cleave both the target and non-target strands (TS & NTS, respectively) three nucleotides upstream of the PAM 82 . In our structure, we observed six NTS nucleotides between the PAM and the scissile phosphate, confirming that PsCas9 introduces double strand breaks as staggered cuts (Supplementary Fig. 1f ), as suggested before 69 .
Collectively, we obtained a high-quality model of PsCas9 RNP, with protein-DNA interaction surfaces and sgRNA folding resolved in exceptional detail. Utilizing these structural insights, along with our biochemical studies, we sought to engineer PsCas9 for enhanced performance in cells via sgRNA scaffold optimization and targeted protein mutagenesis.
Rapid evaluation of Cas9 genome editing activity with luminescence reporter
Amplicon sequencing is a state-of-the-art approach for accurate evaluation of genome editing events. Despite the recent technological advances, NGS-based experimental readout has limited throughput and high cost. To overcome these limitations, we constructed a genome editing luminescence reporter integrated into the genome of HEK293T cells at the HBEGF locus 83 (Supplementary Fig. 2a ). Therein, the Nanoluciferase (Nluc) translation is interrupted by upstream stop codons, which are surrounded by nucleotide sequences with microhomologies. We anticipated that the Cas9-induced DSBs in this locus could undergo repair via the microhomology-mediated end joining (MMEJ) pathway 11 , resulting in the excision of the stop codon, thereby enabling translation of the Nluc gene.
To assess the reporter function and fidelity, we targeted SpCas9 and PsCas9 to the cassette locus with two independent sgRNAs to induce DSBs between the microhomology sites and then analysed the repair outcome with amplicon sequencing. We found that the programmed MMEJ deletion variant was a prevalent repair outcome for both Sp- and PsCas9 (Supplementary Fig. 2b ). We further tested if reporter locus targeting with Cas9 nucleases results in Nluc expression, which we assayed using a luminescence readout. To this end, we overexpressed Sp- or PsCas9 in the reporter cells along with the delivery of corresponding sgRNAs in a fixed amount and later measured luminescence as well as analysed genomic DNA with amplicon sequencing. Background normalized luminescence signal was strongly correlated with the frequency of the MMEJ repair variant, which restores Nluc expression (Supplementary Fig. 2c ). This correlation was confirmed for both Sp- and PsCas9 at a wide range of delivered sgRNA doses highlighting the reliability of our system. As such, this reporter system can be widely employed as an initial screening assay to evaluate the efficacy of compatible Cas9 nucleases without the need for amplicon sequencing analysis, providing faster results and increased throughput.
PsCas9 sgRNA scaffold engineering
Several studies have demonstrated that the activity of various CRISPR-Cas enzymes can be enhanced through the optimization of guide RNA architecture and sequence 67 , 84 , 85 . Our cryo-EM structure exposed an unusual folding of PsCas9’s sgRNA (Fig. 3a, b ). Most characterized Cas9 enzymes contain a sgRNA with a singular, elongated hairpin lying within the REC lobe, formed by repeat and antirepeat sequences stemming from crRNA and tracrRNA, respectively 60 , 62 , 76 , 77 , 78 . Intriguingly, PsCas9’s sgRNA forms a branched structure, where the P1 duplex downstream from the spacer bifurcates into two hairpins, P2 and P3. While the P1 and P3 structures interact with REC1 and Wedge domains and reside within the REC lobe, P2 appears to have minimal contact with the protein (Figs. 2 c, 3a ). Furthermore, the absence of the 10-nucleotide tail downstream of the terminal hairpin P5 in our cryo-EM model suggested its potential redundancy. We hypothesized that the P2 motif and the tail beyond the terminal hairpin could be trimmed without impairing the enzyme’s activity.
a , b sgRNA structure of PsCas9 and its sequence folding. c Editing activity of PsCas9 with different sgRNA scaffold variants analysed using a cell-based luminescence reporter assay. Reporter cells were transfected with plasmid DNA encoding PsCas9 followed by a single dose transfection of a synthetic sgRNA (see Supplementary Fig. 2 ). The luminescence signal of each sgRNA variant was normalized to the initial sgRNA, V2, for both tested targets. Data are shown as mean values for n = 3 biological repeats. d , e Genome editing activity of PsCsa9 combined with original, V1, and optimized, V3, sgRNA scaffold in HEK293T cells mediated via plasmid DNA encoding PsCas9 transfection followed by synthetic RNA transfection at EMX1a ( d ) and PCSK9 ( e ) sites. Data are shown as mean ± SD for n = 3 biological repeats. f , g Structural view of PsCas9 amino acids in the vicinity of the PAM distal DNA end. h , i Evaluation of PsCas9 mutant variants activity using cell-based luminescence reporter. Reporter cells were transfected with plasmid DNA encoding PsCas9 mutants followed by single dose transfection of synthetic sgRNA targeting the cassette. The luminescence signal was normalized to the wild-type, WT, enzyme for both target sites. The screen was performed for a set of single ( h ) and double ( i ) substitution mutant variants. Data are shown as mean values for n = 3 biological repeats. j , k Gene editing activity of PsCsa9 and its engineered E1012R S1314R mutant, ePsCas9, in HEK293T cells mediated via plasmid DNA encoding Cas9 transfection followed by synthetic RNA transfection at EMX1a ( j ) and PCSK9 ( k ) sites. Data are shown as mean ± SD for n = 3 biological repeats.
We therefore designed 15 sgRNA scaffolds with varying levels of P2 hairpin truncation and lacking the 3’- tail (Supplementary Data 2 ). These variants were paired with two spacers targeting the reporter locus and compared with the original sgRNA, which we denote as V1 (Fig. 3c ). Complete excision of the 3’-tail from the sgRNA (V2.1) resulted in ~2-fold increase in editing activity at both target sites. Trimming of the P2 fragment up to its complete removal (V2.2) marginally affected the enzyme’s activity; however, the simultaneous removal of P2 and the 3’-tail resulted in the highest activity variant (V3) with a 2.5-fold increase over V2.
To further validate the performance of the optimized sgRNA scaffold, V3, we targeted two endogenous sites—EMX1a and PCSK9—and measured editing activity with amplicon sequencing, as described earlier (Fig. 3d, e ). At both sites, PsCas9 loaded with V3 sgRNA demonstrated elevated levels of editing across conditions with up to a 2-fold increase over V2 at the highest tested dose. We next investigated whether the in vitro DNA binding of PsCas9 was altered by the optimized sgRNA scaffold. Interestingly, the binding affinity of PsCas9 to the target-containing DNA substrate was improved at both substrates (EMX1a: from ~112 nM to ~5 nM and PCSK9: from ~15 nM to 11 nM) and was in a similar range with SpCas9 (Supplementary Fig. 3a, b ). Interactions with the DNA containing only PAMs but no target site were affected only marginally for both EMX1a and PCSK9 substrates, as evidenced by a negligible increase in polarization signal, suggesting that PsCas9 PAM binding remains markedly weaker than that of SpCas9 (Supplementary Fig. 3c, d ).
Altogether, the engineered sgRNA scaffold of PsCas9 promotes increased genome editing likely through increased binding affinity to the DNA target sequence without affecting PAM interrogation. We hypothesize that trimming of the P2 hairpin may reduce the propensity of the sgRNA to adopt stable alternative, non-native structural conformations, thereby yielding an increased amount of active RNP complex for interaction with DNA both in vitro and inside the cell. This may also be the case for the increased activity observed for the 3’ truncated sgRNA guides.
Rational protein engineering of PsCas9
To further enhance PsCas9 genome editing properties, we designed a set of mutations in the RuvC, Wedge and PI protein domains based on the cryo-EM structure to facilitate the interaction between the PAM proximal DNA tail and the enzyme. Our primary candidates encompassed neutral and negatively charged amino acids predominantly located in flexible regions and proximate to DNA (Fig. 3f, g ). We replaced these with positively charged arginine or polar amino acids to stabilize the protein interaction with the DNA backbone without imposing any sequence specificity (Supplementary Data 3 ).
We introduced plasmids encoding 41 single amino acid substitution mutants of PsCas9 (hereafter called variants) along with a fixed amount of sgRNAs targeting two independent sites to evaluate their relative activity in reporter cells (Fig. 3h ). The control construct, which disrupts the RuvC active site (D10A), led to an almost complete loss of reporter response since Cas9-nickases typically result in minimal genome editing activity. Notably, two tested variants, D947R and S1223R, displayed nickase-like activity levels. D947, a negatively charged residue in proximity to target DNA, fits our initial candidate nomination hypothesis. However, a more careful structural analysis reveals its interaction with R921 and K933 residues in the RuvC domain (Supplementary Fig. 4a ). The D947R substitution might cause strong electrostatic repulsion with these residues, disrupting RuvC folding and its function. Conversely, S1223, located in the DNA proximal surface of the Wedge domain, appears to facilitate PAM recognition through its hydroxyl group’s interaction with the amine of the guanine base at the second position of the PAM (Supplementary Fig. 4b ). Substituting this serine with a bulkier arginine could impair PAM recognition, thereby substantially reducing PsCas9 activity. The majority of other variants either altered the activity marginally or increased it by more than twofold at both targets. Remarkably, two single-substitution variants (E1012R, T1247R) exhibited above 10-fold activity enhancement (Fig. 3h , orange scatters).
We then combined a subset of the activity-enhancing mutants and designed 30 double-substitution variants, which we tested in a similar fashion (Fig. 3i ). As anticipated, several double mutants exhibited further activity improvements, although the effect was not strictly additive, for example with the double E1012R T1247R variant trailing the activity of each single mutant (Supplementary Data 3 ). Conversely, multiple other derivatives of the E1012R variant showed further improvements in editing activity using our reporter assay.
We prioritized a particularly promising variant (E1012R S1314R, named here engineered- or ePsCas9) and assessed its editing properties on endogenous genomic loci. For both tested sites, EMX1a and PCSK9, the editing activity of ePsCas9 was several folds higher compared to the wild-type PsCas9 across all tested sgRNA concentrations (Fig. 3j, k ). At the lower amounts, the activity was improved most noticeably with above 10-fold increase over the wild-type. To assess if the increased genome editing activity of the engineered variant indeed stems from the improved DNA binding properties, we performed further in vitro experiments with recombinant ePsCas9 protein. The affinity of ePsCas9 loaded with sgRNA V3 scaffold was similar to the one of PsCas9 WT at target containing EMX1a and PCSK9 substrates (Supplementary Fig. 3e, f ). While a noticeable increase in the FP signal was observed using EMX1a and PCSK9 substrates containing no target (Supplementary Fig. 3g, h ), it was still markedly lower than that of SpCas9. This finding suggests that PAM interrogation is limiting PsCas9 activity in cells. Collectively, our structure-based rational mutagenesis campaign successfully enhanced PsCas9 genome editing activity introducing ePsCas9.
Our successful efforts in increasing the activity of PsCas9 led us to explore whether this approach could be extended to other Cas9 enzymes, particularly those of the Type II-B subfamily. The structural comparison revealed a high degree of similarity between FnCas9 and PsCas9 (Fig. 2e ). Akin to PsCas9, FnCas9 has been described to have modest genome editing activity in cells 86 and weak DNA interaction in vitro 87 , therefore making it a prime candidate for engineering. We investigated the previously published crystal structure of FnCas9 and selected 12 positions for mutagenesis, applying the same rationale used for PsCas9 (Supplementary Fig. 5a ). Given that FnCas9 shows the same NGG PAM preference, we again employed our cell-based editing assay to evaluate its activity. Wild-type FnCas9 demonstrated reporter activation at both target sites, with a stronger response at target 2 (Supplementary Fig. 5b ). We therefore assessed the performance of the designed FnCas9 variants at target 2. Half of the variants displayed increased activity above 2-folds over the wild-type (Supplementary Fig. 5c ). Three top candidates (E1369R, N1448R and E1603R) underwent further validation across a range of sgRNA concentrations, where they exhibited significant activity enhancements (Supplementary Fig. 5d ). These results indicate that our engineering approach could be applied to other Type II-B family members and enhance their genome editing efficacy.
ePsCas9 is a highly active and specific tool for genome editing
We conducted a thorough evaluation of ePsCas9’s editing properties. Firstly, we picked 18 additional endogenous target sites and compared the on-target editing capabilities of Ps-, ePs-, and SpCas9 using amplicon sequencing (Fig. 4a ). Here, we used a single limiting dose of sgRNAs (0.5 pmol) to avoid editing saturation caused by Cas9 RNP excess. We observed up to 20-fold enhancement in ePsCas9 activity over the enzyme prior engineering, with a median 5-fold increase across tested targets (Fig. 4a and Supplementary Fig. 6 ). ePsCas9 performance was also approaching SpCas9 with comparable median efficacy over the target set (Fig. 4a ). Interestingly, while SpCas9 outperformed ePsCas9 at certain loci (ATTR, B2M2, HEK3), it was outperformed by ePsCas9 at others (B2M1, PCDC1, STAT1), indicating the presence of enzyme-specific target sequence preferences (Supplementary Fig. 6 ). This phenomenon is well characterized in literature 60 , 62 , 66 , 78 with high throughput screening approaches employed to decipher the enzyme-specific sequence preferences 24 , 88 , 89 . Yet, such data set of ~20 targets is insufficient to meaningfully determine ePsCas9’s target selection rules.

a Editing activity of SpCas9, PsCas9 and ePsCas9 across various genomic targets. Editing was induced using plasmid DNA and synthetic sgRNA transfection in HEK293T cells. Distribution of editing at 18 sites for n = 3 biological repeats is represented as combined box-violin plots. The central line shows the median, with the box edges indicating the first (Q1) and third (Q3) quartiles. Whiskers extend to 1.5 times the interquartile range (IQR) from Q1 and Q3, and individual points represent outliers. A two-way ANOVA with Tukey post-hoc test was used to evaluate statistical significance (ns p -value = 0.09, *** p -value < 0.0001; SpCas9-PsCas9 p = 2e-10, ePsCas9-PsCas9 p = 8e-6). b Specificity of SpCas9, PsCas9 and their engineered variants evaluated with CHANGE-seq in the human genome using a promiscuous sgRNA targeting HEKs4 and a specific sgRNA targeting TRAC sites. Specificity values were calculated as the number of on-target reads divided by the total number of reads accounted for by CHANGE-seq hits. Data are shown as mean ± SD for n = 3 technical replicates. c Number of targets consistently discovered across technical replicates of CHANGE-seq. d Translocation frequency induced by SpCas9, PsCas9 and their engineered variants evaluated for two independent events in HEK293T cells. Editing was induced using plasmid DNA and synthetic sgRNAs targeting two loci simultaneously. Translocation frequency was evaluated using ddPCR and normalized to the geometric mean of editing efficiencies at each site. Data are shown as mean ± SD for n = 3 biological repeats. e Schematic of in vivo study to evaluate editing capacity of SpCas9 and ePsCas9 in mice. f in vivo genome editing activity of SpCas9 and ePsCas9. Data are shown as mean ± SD for n = 3, 3 and 4 biological repeats for buffer, SpCas9 and ePsCas9, respectively. g Plasma Pcsk9 levels in mouse plasma post genome editing with SpCas9 and ePsCas9. PCKS9 abundance was evaluated with ELISA at the termination stage. The values were normalized to the mean signal of the buffer treatment group. Data are shown as mean ± SD for n = 3, 3 and 4 biological repeats for buffer, SpCas9 and ePsCas9, respectively. h Correlation between gene editing activity and plasma Pcsk9 level in mice. Pearson correlation value is indicated in the top right corner.
Secondly, we evaluated the performance of ePsCas9 in various cellular contexts. In our previous experiments we employed HEK293T cells, the primary model system for evaluation of gene editors’ performance, due to ease of DNA and sgRNA delivery. Transfection-based plasmid DNA delivery to other cell lines is often inefficient and not tolerated well. To overcome this obstacle, we synthesised ePsCas9 and SpCas9 mRNA. mRNAs with their respective synthetic sgRNAs at a single dose (0.5 pmoles) were transfected to HEK293T cells, as well as to four additional cell lines (HeLa, Huh7, DLD-1 and iPSCs) representing different tissues. We found that at 5 tested target sites editing activity of ePsCas9 remains high and comparable to SpCas9 (Supplementary Fig. 7 ). In some cases, editing was approaching 100% indicating high efficiency of mRNA and sgRNA delivery in cell cultures. Notably, iPSCs displayed the lowest levels of editing across our panel, potentially due to high sensitivity of these cells to transfection procedures in general.
Thirdly, the specificity of ePsCas9 was assessed by analysing its off-target DNA cleavage. Our prior study has demonstrated high fidelity of the wild-type enzyme 69 . However, the increased on-target activity seen in ePsCas9 could potentially suggest an increased activity overall, and thus off-target editing. Hence, we employed CHANGE-seq to examine the off-target activity of Cas9-enzymes in an unbiased manner 90 . We benchmarked ePsCas9 against SpCas9 and its commercially available high-fidelity variant HiFi SpCas9 56 with a well-characterized promiscuous sgRNA targeting the HEKs4 site 43 , as well as with a therapeutically-relevant specific sgRNA targeting the TRAC gene 29 (Fig. 4b ). For all Cas9-RNPs, the on-target sequence was readily detected amongst CHANGE-seq reads while off-target sequences exhibited considerable variation for each enzyme (Supplementary Data 4 and Supplementary Fig. 8a, b ). PsCas9’s specificity was 10- and 5 times higher than that of SpCas9 for HEKs4 and TRAC targets, respectively. While SpCas9 is known to be a promiscuous enzyme, its engineered variant HiFi SpCa9 was also outperformed by PsCas9 several folds (Fig. 4b ). Remarkably, ePsCas9 specificity remained high, comparable to the wild-type enzyme. We also analysed the number of off-targets consistently discovered across technical replicates (Fig. 4c ). For both tested sgRNAs, ePsCas9 showed some increase in identified off-targets compared to the wild-type PsCas9, yet outperformed SpCas9 by at least an order of magnitude and its high fidelity variant HiFi SpCas9 up to 9-fold. Thus, our CHANGE-seq data suggest that the modifications enhancing PsCas9 activity have only a minimal impact on its fidelity.
Lastly, we evaluated the propensity of ePsCas9’s DSBs to promote chromosomal translocations. Large genomic rearrangements including intra- and inter-chromosomal translocations are possible consequences of Cas9 genome editing, and along with off-target editing, raise safety concerns 41 , 42 , 48 . We utilized our previously established translocation assays for two pairs of loci upon simultaneous DNA cuts 69 , 91 . Concurrent DSBs in HEK293T cells were induced using Cas9 nuclease overexpression and delivery of a pair of sgRNAs. The translocation events between the targeted loci were then detected using digital droplet PCR (ddPCR) and normalized to the editing efficiency measured with amplicon sequencing (Fig. 4d ). While SpCas9 displayed high levels of translocations in both assays, translocations induced by PsCas9 were below the detection threshold, potentially due to its low editing activity (Fig. 4d and Supplementary Fig. 9a, b ). ePsCas9 showed high levels of editing at each target and detectable levels of translocations. However, ePsCas9-induced translocations were ten and four times less frequent than that of SpCas9 in HIST1H2BC-HBEGF and PCKS9-HBEGF assays, respectively (Fig. 4d ). Intriguingly, HiFi SpCas9 showed levels of translocations comparable to SpCas9. These findings reiterate that the improved editing capabilities of ePsCas9 only minimally affect its inherent editing fidelity rendering an efficient and safe editor.
In vivo editing by mRNA-encoded ePsCas9 delivered with LNPs
Lipid nanoparticles (LNPs) are recognized for their efficient and transient delivery of genome editing components to the liver and show promise in early clinical trials 30 , 31 , 92 , 93 , 94 . To assess the efficacy of ePsCas9 in therapeutic genome editing with LNP delivery, we focused on the PCSK9 gene, a key target for the treatment of familial hypercholesterolemia (FH) 70 , 71 . PCSK9 disruption through CRISPR-mediated genome editing offers a potentially one-time, long-term therapeutic intervention for FH. Therefore, targeting the Pcsk9 gene in mouse liver provides an ideal model to evaluate the efficacy of genome editing tools. To this end, we formulated LNPs encapsulating ePsCas9 mRNA and sgRNA targeting the mouse Pcsk9 gene using established procedure 95 . We also prepared LNPs containing SpCas9 mRNA and a respective sgRNA with a highly modified scaffold reported previously to facilitate genome editing activity in vivo 92 . These formulations were intravenously injected into C57BL/6NCrl mice, and their editing outcomes was examined in the liver after 7 days (Fig. 4e ).
While SpCas9 with chemically modified sgRNA showed ~ 20% editing, the mice treated with ePsCas9 reached an average of 60% editing, indicating efficient delivery to the mouse liver in vivo (Fig. 4f ). We also analysed Pcsk9 protein levels in blood plasma after LNP delivery and found a reduction in both treated groups compared to the control (Fig. 4g ). As expected, a stronger decrease in Pcsk9 levels correlated with higher levels of genome editing (Fig. 4h ). Furthermore, in mice treated with ePsCas9 as well as SpCas9, we observed no evident safety concerns regarding liver function and overall health condition (Supplementary Fig. 10a, b ), as evaluated by body weight monitoring and liver enzymes activity. Collectively, these findings suggest that ePsCas9 is an efficient and safe editor for in vivo genome editing when delivered using LNPs.
The advent of CRISPR editing technologies made genome modifications in vivo accessible. However, a key challenge in therapeutic genome editing lies in the efficient delivery of CRISPR components 33 . LNPs have recently emerged as a viable delivery modality for the liver, due to their cost-effectiveness and improved safety profile compared to viral vectors 92 , 93 , 94 . However, the swift metabolism and limited amount of RNA payloads present a significant obstacle, narrowing the window for efficacious genome editing.
Our previous work introduced PsCas9, a Type II-B family enzyme that demonstrated high fidelity and notable activity in vivo when delivered via viral vectors 69 . However, in a scenario mimicking LNP delivery within cell cultures, PsCas9 displayed limited editing activity (Fig. 1 ).
In this study, we leveraged the high-resolution cryo-EM structure of PsCas9 to guide enzyme engineering (Fig. 2 ). We hypothesized that PsCas9’s modest editing activity could be attributed to its limited DNA-binding ability in vitro (Fig. 1 ). Through a combination of protein and sgRNA modifications (Fig. 3 ), we achieved a boost in DNA binding and substantially improved PsCas9’s editing efficacy in cell cultures across multiple targets and cellular contexts. We also showed engineered PsCas9 applicability for therapeutically relevant in vivo genome editing (Fig. 4 ). At the single tested Pcsk9 target, ePsCas9 induces a higher level of editing than SpCas9 in the liver and a concurrent decrease of Pcsk9 levels in blood plasma. More in vivo experiments will be required to compare the performance of both nucleases at other loci.
Our mutagenesis efforts improved PsCas9 PAM interrogation activity modestly yet that was sufficient to enhance its editing activity up to 20-fold at some target sites. This suggests that PsCas9 initial DNA engagement is the rate limiting step for catalysing DNA cleavage in cellulo . Substantial enhancement in PsCas9 editing activity was achieved by introducing positively charged amino acids to DNA-interacting domains. It has been established that the NGG PAM recognition is a weak interaction, with an estimated dissociation constant of ~10 µM 96 . We propose that the E1012R E1314R and T1247R mutations contribute the enhanced DNA binding affinity of ePsCas9 through introduction of additional non-specific electrostatic contacts with the DNA phosphodiester backbone. This is reminiscent of the enhanced affinity of the PAMless variant SpRY-Cas9 which also uses non-specific contacts to enhance DNA binding affinity 97 . We successfully applied this approach to improve the editing efficacy of FnCas9, an enzyme phylogenetically relevant to PsCas9 albeit sharing only ~20% sequence homology. We believe this strategy could be further generalized and applied for the engineering of a broader range of RNA-guided nucleases in the Type II family.
In the future, it might be possible to add the SpRY or SpG mutations (or establish equivalent mutations that could be introduced) to ePsCas9 to broaden the DNA targeting abilities. However, one must consider that SpG and SpRY have ~25- and ~500-fold reduction in DNA cleavage rates relative to SpCas9 97 , and given that such mutations are also likely to severely impact the on-target DNA cleavage efficiency of ePsCas9 it is worth evaluating how necessary PAM-flexible DNA targeting is for the desired genome editing application.
The therapeutic application of CRISPR-based genome editing raises multiple safety concerns 38 , 39 , 40 , 41 , 42 , 43 , 44 , 45 , 46 , 47 , 48 , 49 , 50 . Editing at off-target sites that share a high level of homology with the intended target is a well-documented phenomenon potentially introducing harmful unwanted mutations. Off-target editing is mitigated by the introduction of high-fidelity Cas-enzyme through rational engineering, directed evolution and natural variants discovery 51 , 52 , 53 , 54 , 55 , 56 , 57 , 58 , 59 . Biochemical studies 74 , 98 proposed that differences in Cas9 RNP dissociation rates between perfect and imperfect substrates are the major determinants for the DNA cleavage, and thus, off-target editing. In our previous work, we introduced PsCas9 as an enzyme with exceptional fidelity and virtually no off-target editing in vivo, which we attributed to its strong discrimination in off- vs on-target DNA cleavage in vitro 69 . Here, we boosted PsCas9 activity while preserving its high biochemical fidelity and favourable off-target profile as demonstrated by only a minor alteration of the enzyme’s specificity measured by CHANGE-seq.
Large genomic rearrangements, including chromosomal translocations, have been recently identified as an on-target consequence of Cas9-based genome editing that pose an additional safety risk 41 , 45 , 48 , 49 . Moreover, multiplexed editing introduces the potential for translocations between simultaneously targeted sites. Here, we showed that engineered PsCas9 introduces fewer translocations than SpCas9, maintaining the properties of the wild-type enzyme 69 . We also observed that the high-fidelity SpCas9 variant, HiFi SpCas9, introduces high levels of genomic translocations, despite its higher fidelity in off-target editing 56 . We believe that the low translocation properties of Ps- and ePsCas9 could be attributed to their specific DNA cleavage pattern forming 5’-overhangs. Well documented observations of Cas12-family enzymes to induce fewer translocations and generate staggered DSBs further support our hypothesis. 61 , 67 , 68 , 99 . Non-matching sticky DNA ends at DSB potentially pose a bigger challenge to repair for NHEJ machinery than blunt ends, and thus, inhibit translocation formation. In this context, ePsCas9 can serve as a safer alternative to existing editing tools with high on-target and low off-target activity providing the advantage of fewer translocations, in particular for multiplexed editing.
Overall, our study presents an additional tool for the CRISPR toolbox: a high-fidelity and high-activity genome editor, which is effective for in vivo applications using LNP delivery. While SpCas9 remains the gold standard in the field due to its remarkable activity, versatility, and wealth of accumulated research, several recently introduced genome editors offer competitive activity and enhanced fidelity. Even though these emerging enzymes may perform comparably on average, specific genomic locations may be better suited to one enzyme over another. Thus, the expansion of the CRISPR toolbox with additional enzymes greatly enhances our capacity to develop efficient genome editing therapies for a wider range of targets.
Ethical statement
All mouse experiments were approved by the AstraZeneca internal committee for animal studies and the Gothenburg Ethics Committee for Experimental Animals (license numbers: 162-2015+ and 2194-2019) compliant with EU directives on the protection of animals used for scientific purpose.
Cell culture and transfection procedures
HEK293T (GenHunter Corporation, Q401), HeLa (ATCC, CCL-2) and Huh7 (Riken Cell bank, RCB1366) cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% foetal bovine serum (FBS). DLD-1 (ATCC, CCL-221) cells were cultured in RPMI 1640 + 2 mM Glutamine + 10%FBS. hiPSC were generated and maintained in a feeder-free human pluripotency culturing system, Cellartis DEF-CS 500 (Takara, Japan), according to manufacturer’s instructions 100 . The cells were maintained at 37 °C in a humidified incubator with 5% CO 2 . For transfection experiments, cells were seeded at a density of 2 × 10^4 cells per well in 96-well plates, 24 h prior to transfection.
For plasmid-based sgRNA delivery, cells were transfected using a mixture of 40 ng and 80 ng of plasmids encoding Cas9 and sgRNA, respectively. The transfection was carried out using 0.3 µl of FuGene reagent (Promega) in a final volume of 5 µl.
In experiments involving synthetic sgRNA delivery, 50 ng of the Cas9-encoding plasmid was initially transfected using 0.3 µl of FuGene reagent (Promega) in a final volume of 5 µl. After a 24 h interval, varying quantities of synthetic sgRNA (Synthego or Integrated DNA technologies, IDT) were transfected into the cells using 0.5 µl of Lipofectamine RNAiMAX reagent (Thermo Fisher Scientific) in a final volume of 10 µl. Plasmid sequences are provided in Supplementary Data 5 .
In experiments involving simultaneous mRNA and sgRNA transfection, 100 ng of mRNA and 0.5 pmoles of respective sgRNA were transfected using 0.3 µl of Lipofectamine MessengerMAX (Thermo Fisher Scientific) in a final volume of 10 µl.
Amplicon Sequencing and editing efficiency analysis
To assess genome editing efficacy in HEK293T cells, genomic DNA was isolated 72 h post-transfection using the QuickExtract DNA Extraction Solution (Lucigen), following the manufacturer’s instructions in 50 ul final volume.
Primary amplicons were synthesized using the Phusion Flash High-Fidelity PCR Master Mix (Thermo Fisher Scientific). Reactions were set up in a 15 µl volume, comprising 0.25 µM target-specific primers (IDT) and 1.5 µl of genomic DNA. The PCR cycling conditions were set as follows: initial denaturation at 98 °C for 1 min, followed by 32 cycles of 98 °C for 10 s, 60–65 °C for 10 s, and 72 °C for 10 s. Post amplification, PCR products were cleaned up using Ampure XP beads (Beckman Coulter) and analysed on a Fragment Analyzer (Agilent Technologies).
For the indexing step, a secondary PCR was conducted using KAPA HiFi HotStart Ready Mix (Roche). The reaction included 1 ng of the primary amplicon and 0.5 µM of indexing primers (IDT) in a 25 µl total reaction volume. The thermal cycling conditions included an initial 72 °C for 3 min, followed by 98 °C for 30 s, then 10 cycles of 98 °C for 10 s, 63 °C for 30 s, and 72 °C for 3 min; followed by a final extension at 72 °C for 5 min. Post-indexing, amplicons were again purified using Ampure XP beads and quality controled on the Fragment Analyzer.
For sequencing library quantification, the Qubit 4 Fluorometer (Thermo Fisher Scientific) was employed. High-throughput sequencing was executed on the Illumina NextSeq platform, in adherence to the manufacturer’s guidelines. All primers, amplicon reference sequences, and target sites utilized in this study are catalogued in Supplementary Data 6 , 7 .
NGS data was demultiplexed by using bcl2fastq software. The fastq files were analyzed by CRISPResso 101 version 2.2.12 with the following parameters: -q 30 –ignore substitutions max_paired_end_reads_overlap 300 -w 15 -wc −3.
Translocations assay
Translocation frequency between two simultaneously targeted sites was evaluated using ddPCR and amplicon sequencing 69 , 91 . Briefly, balanced translocations between either HIST1H2BC-HBEGF or PCSK9-HBEGF were detected using custom FAM-labelled ddPCR assays (Bio-Rad). HEX-labelled AP3B1 assay (Bio-Rad, dHsaCP1000001) was used as reference. Sequences for the primers and probes are listed in Supplementary Data 7 .
20 µl ddPCR reaction mixes were prepared, each containing 1x ddPCR Supermix for Probes (no UTP) (Bio-Rad), 1x FAM-labelled custom translocation assay, 1x HEX-labelled reference assay, 1/40 HaeIII (NEB), 5 µl of 1:5 diluted QuickExtract DNA solution and ultrapure RNase- and DNase-free water (Invitrogen). An automated Droplet Generator (Bio-Rad) was used to generate droplets and C1000 Touch Thermal Cycler (Bio-Rad) for PCR amplification. Following PCR conditions were used: 95 °C for 10 min, followed by 40 cycles of 94 °C for 30 s and 61/63 °C for 1 min, followed by 98 °C for 10 min. All steps were performed with ramp rate fixed at 2 °C/s.
Droplet reading was performed with the QX 200 Droplet reader (Bio-Rad) using ddPCR Droplet Reader Oil (Bio-Rad). The QX Manager (Bio-Rad) software was used for data acquisition and analysis. The fluorescence amplitude threshold was set manually, using the midpoint between the average fluorescence amplitude of the FAM and HEX channels in positive samples and the negative control. The same threshold was applied to all wells of the ddPCR-plate using the same translocation assay. Editing efficiencies at the individual target sites were quantified using amplicon sequencing. Normalized translocation frequency was evaluated as observed frequency from ddPCR divided by geometric mean of editing efficiencies at each site.
Cell-based genome editing reporter assay
The genome editing reporter was developed employing the previously described Xential method 83 . In summary, HEK293T cells, at a density of 0.5 × 10 ^ 6 cells/well, were seeded onto 6-well plates. These cells were transfected with plasmid DNAs encoding Cas9, an sgRNA targeting intron 3 of the HBEGF gene, and a repair template incorporating the reporter cassette. Transfections were facilitated by the FuGene reagent (Promega) with the total transfection mixture of 150 µl, comprising 3 µg of total DNA with relative ratio of 1:1:2 (Cas9:sgRNA:repair template). A total of 9 µl of the FuGene reagent was used for each well. Seventy-two hours post-transfection, the cells underwent selection using diphtheria toxin (DT) at final concentration of 10 ng/ml for a duration of 1 week. Following this, cells were transitioned to a DT-free medium and allowed to expand.
For reporter activation, the cells were transfected with plasmid DNA encoding Cas9s and synthetic sgRNA designed to target the reporter cassette. The same transfection procedure described in “cell culture and transfection procedures” section was followed. After 72 h, the culture medium was replaced with fresh warm medium. Six to twelve hours later, the medium was harvested for luminescence assessment. 10 µl of the collected medium was mixed with 10 µl of Nano-Glo® Luciferase substrate (Promega) diluted at 1:1000 in PBS. Luminescence measurements were carried out on the PheraStar FSX plate reader (BMG Labtech). To test reporter fidelity and performance, the cassette locus was also analysed with amplicon sequencing as described above.
Modified CHANGE-seq
CHANGE-seq was performed as previously described by Lazzarotto et al. 90 with additional modifications. As opposed to SpCas9, PsCas9 generates staggered ends, impeding the ligation of the sequencing adaptor to the DNA double stranded breaks (DSBs). Thus, we introduced a DNA blunting step prior to adaptor ligation, as described in our previous work 69 . Here, we introduced an additional end-repair step in the presence of ddNTPs prior to Cas9 cleavage to block non-specific DNA ends for adaptor ligation, and thus, reduce the background noise reads.
High Molecular weight genomic DNA (Promega) was subjected to tagmentation with a custom Tn5-transposome harbouring oCRL225/oCRL226 adaptors and the Hyperactive Tn5 Transposase (Diagenode). DNA tagmentation was performed in batches of 2 µg, utilizing 8.7 µl of the assembled transposome in a final volume of 200 µl of 1x Tagmentation Buffer (Diagenode) and incubated for 7 minutes at 55 °C. Reaction was quenched by the addition of 200 µl of SDS 0.4%, and resultant fragments were assessed on the Fragment analyzer and quantified by Qubit dsDNA BR Assay kit (Thermo Fisher Scientific). Tagmented DNA was then subjected to gap repair with Kapa Hi-Fi HotStart Uracil+ DNA Polymerase (KAPA Biosystems) and Taq DNA Ligase (NEB). Resultant gap-repaired DNA was treated with USER enzyme (NEB) and T4 polynucleotide kinase (NEB), and then circularized overnight with T4 DNA Ligase (NEB) and treated with a cocktail of exonucleases containing Plasmid-Safe ATP-dependent DNase (Lucigen), Lambda exonuclease (NEB) and Exonuclease I (NEB) to degrade residual linear DNA carryover. To avoid capturing pre-existing non-specific dsDNA breaks at the end-repair step, exonuclease-treated circles were then further subjected to 3’end blocking by incubating 160 ng circularized material with 7.5 U of T4 DNA Polymerase, 7.5 U of Klenow Fragment (3’ → 5’ exo-), in presence of 0.1 mM of ddNTP, in a final volume of 100 ul of 1x T4 DNA Ligation Buffer. The mixture was incubated at 20 degrees for 30 minutes and bead purified with AmpureXP beads at 1:1 ratio. 150 ng of circularized material were in vitro cleaved by SpCas9, HiFi SpCas9, wild-type PsCas9 and ePsCas9 RNPs in combination with HEK4 and TRAC sgRNAs (sequences details in Supplementary Data 7 ) in a total volume of 50 µl.
All libraries were subjected to an end-repair step using T4 DNA polymerase (NEB), so that 5‘ overhangs are filled to form blunt ends for ligation. Then, Illumina Universal Adaptor (NEB) was ligated to adenylated blunt ends, enzymatically treated with USER enzyme (NEB) and amplified with NEBNext Multiplex Oligos for Illumina for 20 amplification cycles. The quality of the amplified and bead-cleaned-up libraries was determined using a 5300 Fragment analyzer with the standard sensitivity NGS kit (Agilent). Libraries were further quantified by qPCR (Thermo Fisher Scientific), pooled and denatured according to Illumina’s recommendations and sequenced on a NextSeq550 on a PE150 configuration, to achieve a mean coverage of ~16 M reads per library. The sequenced reads were analyzed using the published version of the CHANGE-seq pipeline 90 with minor modifications. The pipeline was run with the following parameters: read_threshold: 4, window_size: 3, mapq_threshold: 50, start_threshold: 1, gap_threshold: 3, mismatch_threshold: 6, search_radius: 30, merged_analysis: False, PAM = NNN. Reads with MAPQ = 0 were included in the analysis alongside those passing the MAPQ threshold defined in the parameters, to nominate putative off-targets located in non-uniquely mappable regions.
Purification of Cas9 proteins
While SpCas9 and HiFi SpCas9 proteins were procured from IDT, the PsCas9 and ePsCas9 proteins were purified using a previously established protocol 69 . Briefly, the E. coli BL21 λDE3 star strain was transformed with pET24a-based expression vectors. Freshly transformed colonies were cultivated overnight in LB medium, then sub-cultured into 800 ml of TB medium. This culture was maintained at 37 °C until an optical density OD600 approached ~2, under robust agitation. The growth temperature was subsequently reduced to 18 °C, and after a 1-h, isopropyl β-D-1-thiogalactopyranoside (IPTG) was introduced to a final concentration of 200 µM to induce protein expression. Post an overnight incubation, cells were harvested via centrifugation.
Cell lysis was achieved through high-pressure disintegration in a buffer comprising 20 mM HEPES (pH 7.5), 150 mM KCl, 5% glycerol, and 1 mM dithiothreitol (DTT). The resulting lysate was clarified by centrifugation and then subjected to affinity chromatography using a 5 ml HisTrap column (Cytiva). After equilibrating the column with a buffer containing 20 mM imidazole, bound proteins were eluted using a buffer with 300 mM imidazole. Relevant protein fractions were further subjected to gel filtration using a Superdex 200 10/600 column (Cytiva), pre-equilibrated with a buffer of 20 mM HEPES (pH 7.5), 300 mM NaCl, 5% glycerol, and 1 mM DTT. The purified Cas9 protein fractions were then concentrated to a final concentration of 10 mg/ml, flash frozen using liquid nitrogen, and stored at −80 °C until further use. The sequences of utilized plasmids can be found in Supplementary Data 5 .
DNA binding assay using fluorescence polarization
For the DNA binding assay, we employed a fluorescence polarization technique, adapting a method that has been previously described 75 . FAM-labelled DNA oligonucleotides were procured from IDT. To prepare dsDNA substrates, the oligos were annealed at a concentration of 10 µM in an annealing buffer composed of 10 mM TRIS-HCl (pH 7.5) and 50 mM KCl. The solution was heated at 95 °C for 5 min and allowed to cool down gradually to room temperature. Once annealed, the dsDNA was diluted to a final concentration of 20 nM using a binding buffer containing 20 mM TRIS-HCl, 200 mM KCl, 5% Glycerol, and 10 mM CaCl2.
sgRNAs were refolded using a similar procedure, with an initial concentration of 10 µM in the annealing buffer. To form the Cas9 ribonucleoproteins (RNPs), Cas9 protein and its respective sgRNA were mixed at concentrations of 2 µM and 2.5 µM, respectively, in the binding buffer. This mixture was then incubated at room temperature (~25 °C) for 20 min. The Cas9 RNPs were subsequently serially diluted using the binding buffer. Equal volumes of the diluted RNPs and dsDNA substrates were combined, resulting in a final concentration of 10 nM DNA. The reactions were allowed to incubate at room temperature for an additional 15 minutes. Fluorescence polarization reading of FAM fluorophore was then taken using the PheraStar FSX plate reader (BMG Labtech). The sequences of utilized oligonucleotides are provided in Supplementary Data 7 .
Cryo-EM data processing
PsCas9 RNP complex rapidly thawed and incubated with 4-fold excess of heat-annealed (90 °C for 5 min, and rapidly cooled to 4 °C) EMX1a dsDNA, and incubated at room temperature (~25 °C) for 30 min prior to vitrification. 2.5 µl of this complex were applied to C-flat holey carbon grids (1.2/1.3, 300 mesh), which had been plasma cleaned for 30 s in a Solarus 950 plasma cleaner (Gatan) with a 4:1 ratio of O 2 /H 2 . Grids were blotted with Vitrobot Mark IV (Thermo Fisher Scientific) for 2 s, blot force 4 at 4 °C & 100% humidty, and plunge-frozen in liquid ethane. Data were collected using a FEI Titan Krios cryo-electron microscope equipped with a K3 Summit direct electron detector (Gatan, Pleasanton, CA). Since initial data processing revealed a severe preferred orientation, the full dataset was collected with the stage tilted at −30°. Images were recorded with SerialEM 102 with a pixel size of 0.81 Å. A total accumulated dose of 70 electrons/Å 2 during a 6 s exposure was fractionated into 80 frames, at a defocus range of −1.5 to −2.5 µm. A total of 7938 micrographs were collected, of which 6041 with CTF fits of 5 Å or better were retained. Motion correction, CTF estimation and particle picking was performed on-the-fly using cryoSPARC Live v4.0.0-privatebeta.2 103 . All subsequent data processing was performed in cryoSPARC v3.2 104 . Data processing workflow is provided in Supplementary Fig. 11 .
A total of 3,932,646 particles were picked, of which 773,096 were selected after 2D classification. Multiple rounds of ab initio reconstruction and heterogeneous refinement resulted in a subset of 433,192 particles which used for a consensus reconstruction, which was resolved to 3.0 Å-resolution using non-uniform refinement. This subset of particles was then further classified using he 3D classification job within cryoSPARC ( k = 10). One class of particles was well-resolved and significantly more abundant than the other classes (81,473 particles), which was then used for subsequent non-uniform refinement. After multiple rounds of CTF refinement, a 2.86 Å resolution reconstruction was determined. This map was then used for modelling.
Model building and figure preparation
An AlphaFold2 model of PsCas9 was generated, and individual domains were rigid body fitted into the unsharped reconstruction. Once all protein density had been accounted for, the individual domains were connected, and the nucleic acid chains were build de novo in Coot 105 . Once fully modelled, Isolde 106 was used to improve the fit of the model to the map, and real-space refinement as implemented within Phenix 107 was performed to optimize model geometry. All structural figures and movies were generated using ChimeraX 108 , 109 .
mRNA synthesis
mRNA is synthesised by a T7 polymerase driven in vitro transcription (IVT) reaction from a plasmid DNA template which contains a T7 polymerase promoter sequence upstream of all the elements required in the mRNA. mRNA comprises a 5’ cap structure incorporated through the inclusion of a cap analogue (CleanCap AG®) during mRNA synthesis; a 5’ untranslated region sequence; a nuclear localisation sequence genetically fused to the coding sequence of Cas9 followed by another nuclear localisation sequence; a 3’ untranslated region sequence and a defined polyA tail (80–120 bp). The mRNA is prepared with the replacement of uridine by N1-Methyl-Pseudouridine or 5-Methoxyuridine to minimize recognition of the IVT produced mRNA by the innate immune system. The DNA template is linearized downstream of the polyA tail by BspQI restriction endonuclease before IVT to ensure all mRNA molecules terminate directly after the polyA tail.
LNP Formulation for In Vivo Cas9 delivery
Lipid nanoparticles (LNPs) were synthesized in line the with previously established protocols 95 . Specific lipids were solubilized in ethanol at final concentration of 12.5 mM. Separately, Cas9 mRNA and corresponding sgRNAs (Axolabs) were diluted in RNase-free 50 mM citrate buffer (pH 3.0). The ethanol-based lipid solution and the aqueous mRNA/sgRNA solution were then combined at a 3:1 volume ratio utilizing the microfluidic NanoAssemblr Ignite device (Precision NanoSystems) with a set mixing flow rate of 12 mL/min. The resultant LNPs underwent an overnight dialysis using Slide-A-Lyzer G2 10 K MWCO dialysis cassettes (Thermo Fisher Scientific) in PBS, pH 7.4.
Particle size distribution and overall size were determined via dynamic light scattering (DLS) with the Zetasizer Nano-ZS instrument (Malvern Instruments). The formulated LNPs were between 70 and 78 nm in size, with a polydispersity index (PDI) in the range of 0.05 to 0.07. The LNPs were concentrated using Amicon Ultra 4 30,000 kDa MWCO centrifugal filter units, and then DLS was performed again to ascertain LNP structural integrity. To quantify the encapsulated RNA within the LNPs, the RiboGreen assay (Thermo Fisher Scientific) was utilized. The encapsulation efficiency was above 90% for all LNP samples. LNPs were then diluted to the working RNA concentration of 0.2 mg/ml.
In vivo LNP delivery
Female C57Bl/6NCrl mice were acquired from Charles River Laboratories. Animals were maintained in a controlled environment with a room temperature of 21 °C, a relative humidity ranging between 45–55%, and a 12:12 h light-dark cycle (lights on at 6:00 am, lights off at 6:00 pm). Throughout the study, mice were granted unrestricted access to a standard chow diet (R70, Lactamin AB, Stockholm, Sweden) and water. Enrichment elements, such as cardboard tubes, chew sticks, and shredded paper, were provided in the cages. Animals underwent daily health inspections and were weighed weekly.
Mice, aged between 10–12 weeks, were administered LNPs at a dose of 1 mg/kg via lateral tail vein injection. As a comparison, a control group was injected with an equivalent volume of buffer. One week following the injections, mice were euthanized, and liver tissues collected for analysis. The left median liver lobes were designated for genomic DNA extraction using the Puregene Tissue Kit (Qiagen). Subsequent analyses to determine genome editing efficiencies were executed via amplicon sequencing, as detailed in the prior section. Animal sex was not considered in the study due to the use of sex-independent genomic locus investigated.
Assessment of liver function following LNP delivery
To gauge potential hepatic impact post-LNP administration, plasma levels of alanine transaminase (ALT) and aspartate aminotransferase (AST) were assessed. Blood was collected at termination via retroorbital eye bleed. Blood samples of ~600 μL were drawn into 500 LiHep Microvette tubes. Plasma was separated from the blood within 30 min of collection using centrifugation at 1500 g for 10 minutes at 4 °C. Resultant plasma samples were preserved at −20 °C until dispatched to Charles River Laboratories (Edinburgh, UK), where the specific activities of ALT and AST were determined.
Statistics and reproducibility
No statistical method was used to predetermine sample size. Sample sizes for in vitro and in cellulo experiments were selected based on literature precedence for genome editing experiments. No data was excluded. All cell and in vitro experiment were independently repeated at least once as specified in figure legends. Mammalian cells were cultured under identical conditions, no randomization was used. Animal experiments: Three to four animals were included per group. Animals were randomized based on their weights measured prior to the experiments. The investigators were not blinded to allocation during experiments and outcome assessment. Statistical tests used described in the figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Source data are provided as a Source Data file. The structure of PsCas9 and its associated atomic coordinates, has been deposited into the EMDB and the PDB repositories with EMDB accession number EMDB-42378 and PDB accession number 8UMF , respectively. NGS datasets generated in this study are deposited to the NIH Sequence Read Archive with BioProject accession numbers: PRJNA1154610 and PRJNA1154611 . Previously reporter FnCas9 protein structure used in this work (PDB accession number 5B2O ). All the sequences of sgRNAs, mRNAs, proteins and plasmids used in the study are available in the Supplementary Data file. Source data are provided with this paper. Synthetic guide RNAs and ePsCas9 are available from Synthego.
Code availability
No specific code was developed in regard to this publication.
Barrangou, R. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315 , 1709–1712 (2007).
Article ADS CAS PubMed Google Scholar
Brouns, S. J. et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321 , 960–964 (2008).
Article ADS CAS PubMed PubMed Central Google Scholar
Al-Shayeb, B. et al. Clades of huge phages from across earth’s ecosystems. Nature 578 , 425–431 (2020).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337 , 816–821 (2012).
Cho, S. W., Kim, S., Kim, J. M. & Kim, J. S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31 , 230–232 (2013).
Article CAS PubMed Google Scholar
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339 , 819–823 (2013).
Jinek, M. et al. RNA-programmed genome editing in human cells. Elife 2 , e00471 (2013).
Article PubMed PubMed Central Google Scholar
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339 , 823–826 (2013).
Bibikova, M., Golic, M., Golic, K. G. & Carroll, D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics 161 , 1169–1175 (2002).
Article CAS PubMed PubMed Central Google Scholar
Bibikova, M., Beumer, K., Trautman, J. K. & Carroll, D. Enhancing gene targeting with designed zinc finger nucleases. Science 300 , 764 (2003).
van Overbeek, M. et al. DNA repair profiling reveals nonrandom outcomes at Cas9-mediated breaks. Mol. Cell 63 , 633–646 (2016).
Article PubMed Google Scholar
Bibikova, M. et al. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol. Cell Biol. 21 , 289–297 (2001).
Chen, F. et al. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat. Methods 8 , 753–755 (2011).
Article MathSciNet CAS PubMed PubMed Central Google Scholar
Ran, F. A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8 , 2281–2308 (2013).
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533 , 420–424 (2016).
Gaudelli, N. M. et al. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 551 , 464–471 (2017).
Anzalone, A. V. et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576 , 149–157 (2019).
Maeder, M. L. et al. CRISPR RNA-guided activation of endogenous human genes. Nat. Methods 10 , 977–979 (2013).
Mali, P. et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 31 , 833–838 (2013).
Qi, L. S. et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152 , 1173–1183 (2013).
Platt, R. J. et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159 , 440–455 (2014).
Swiech, L. et al. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat. Biotechnol. 33 , 102–106 (2015).
Yang, H. et al. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154 , 1370–1379 (2013).
Shalem, O. et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343 , 84–87 (2014).
Wang, T., Wei, J. J., Sabatini, D. M. & Lander, E. S. Genetic screens in human cells using the CRISPR-Cas9 system. Science 343 , 80–84 (2014).
Zhou, Y. et al. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature 509 , 487–491 (2014).
Liu, X. et al. CRISPR-Cas9-mediated multiplex gene editing in CAR-T cells. Cell Res 27 , 154–157 (2017).
Article ADS PubMed Google Scholar
Rupp, L. J. et al. CRISPR/Cas9-mediated PD-1 disruption enhances anti-tumor efficacy of human chimeric antigen receptor T cells. Sci. Rep. 7 , 737 (2017).
Article ADS PubMed PubMed Central Google Scholar
Eyquem, J. et al. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature 543 , 113–117 (2017).
Frangoul, H., Ho, T. W. & Corbacioglu, S. CRISPR-Cas9 gene editing for sickle cell disease and beta-thalassemia reply. N. Engl. J. Med 384 , e91 (2021).
Gillmore, J. D. et al. CRISPR-Cas9 in vivo gene editing for transthyretin amyloidosis. N. Engl. J. Med. 385 , 493–502 (2021).
Koonin, E. V. & Makarova, K. S. Evolutionary plasticity and functional versatility of CRISPR systems. PLoS Biol. 20 , e3001481 (2022).
Wang, J. Y. & Doudna, J. A. CRISPR technology: a decade of genome editing is only the beginning. Science 379 , eadd8643 (2023).
Saito, M. et al. Fanzor is a eukaryotic programmable RNA-guided endonuclease. Nature 620 , 660–668 (2023).
Karvelis, T. et al. Transposon-associated TnpB is a programmable RNA-guided DNA endonuclease. Nature 599 , 692–696 (2021).
Altae-Tran, H. et al. The widespread IS200/IS605 transposon family encodes diverse programmable RNA-guided endonucleases. Science 374 , 57–65 (2021).
FDA. CASGEVY. https://www.fda.gov/vaccines-blood-biologics/casgevy (2022).
Fu, Y. et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 31 , 822–826 (2013).
Hsu, P. D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31 , 827–832 (2013).
Pattanayak, V. et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 31 , 839–843 (2013).
Ghezraoui, H. et al. Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. Mol. Cell 55 , 829–842 (2014).
Frock, R. L. et al. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat. Biotechnol. 33 , 179–186 (2015).
Tsai, S. Q. et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 33 , 187–197 (2015).
Shin, H. Y. et al. CRISPR/Cas9 targeting events cause complex deletions and insertions at 17 sites in the mouse genome. Nat. Commun. 8 , 15464 (2017).
Kosicki, M., Tomberg, K. & Bradley, A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 36 , 765–771 (2018).
Cullot, G. et al. CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations. Nat. Commun. 10 , 1136 (2019).
Alanis-Lobato, G. et al. Frequent loss of heterozygosity in CRISPR-Cas9-edited early human embryos. Proc. Natl Acad. Sci. USA 118 , e2004832117 (2021).
Leibowitz, M. L. et al. Chromothripsis as an on-target consequence of CRISPR-Cas9 genome editing. Nat. Genet 53 , 895–905 (2021).
Papathanasiou, S. et al. Whole chromosome loss and genomic instability in mouse embryos after CRISPR-Cas9 genome editing. Nat. Commun. 12 , 5855 (2021).
Hoijer, I. et al. CRISPR-Cas9 induces large structural variants at on-target and off-target sites in vivo that segregate across generations. Nat. Commun. 13 , 627 (2022).
Kleinstiver, B. P. et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529 , 490–495 (2016).
Slaymaker, I. M. et al. Rationally engineered Cas9 nucleases with improved specificity. Science 351 , 84–88 (2016).
Chen, J. S. et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 550 , 407–410 (2017).
Casini, A. et al. A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nat. Biotechnol. 36 , 265–271 (2018).
Lee, J. K. et al. Directed evolution of CRISPR-Cas9 to increase its specificity. Nat. Commun. 9 , 3048 (2018).
Vakulskas, C. A. et al. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat. Med 24 , 1216–1224 (2018).
Schmid-Burgk, J. L. et al. Highly parallel profiling of Cas9 variant specificity. Mol. Cell 78 , 794–800.e798 (2020).
Bravo, J. P. K. et al. Structural basis for mismatch surveillance by CRISPR-Cas9. Nature 603 , 343–347 (2022).
Kim, Y. H. et al. Sniper2L is a high-fidelity Cas9 variant with high activity. Nat. Chem. Biol. 19 , 972–980 (2023).
Nishimasu, H. et al. Crystal structure of staphylococcus aureus Cas9. Cell 162 , 1113–1126 (2015).
Zetsche, B. et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163 , 759–771 (2015).
Hirano, H. et al. Structure and engineering of francisella novicida Cas9. Cell 164 , 950–961 (2016).
Kim, E. et al. In vivo genome editing with a small Cas9 orthologue derived from campylobacter jejuni. Nat. Commun. 8 , 14500 (2017).
Edraki, A. et al. A compact, high-accuracy Cas9 with a dinucleotide PAM for in vivo genome editing. Mol. Cell 73 , 714–726 e714 (2019).
Kleinstiver, B. P. et al. Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat. Biotechnol. 37 , 276–282 (2019).
Schmidt, M. J. et al. Improved CRISPR genome editing using small highly active and specific engineered RNA-guided nucleases. Nat. Commun. 12 , 4219 (2021).
Kim, D. Y. et al. Efficient CRISPR editing with a hypercompact Cas12f1 and engineered guide RNAs delivered by adeno-associated virus. Nat. Biotechnol. 40 , 94–102 (2022).
Hino, T. et al. An AsCas12f-based compact genome-editing tool derived by deep mutational scanning and structural analysis. Cell 186 , 4920–4935.e23 (2023).
Bestas, B. et al. A Type II-B Cas9 nuclease with minimized off-targets and reduced chromosomal translocations in vivo. Nat. Commun. 14 , 5474 (2023).
Katzmann, J. L., Cupido, A. J. & Laufs, U. Gene therapy targeting PCSK9. Metabolites 12 , 70 (2022).
Ding, Q. et al. Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ. Res 115 , 488–492 (2014).
Sternberg, S. H., Redding, S., Jinek, M., Greene, E. C. & Doudna, J. A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507 , 62–67 (2014).
Dagdas, Y. S., Chen, J. S., Sternberg, S. H., Doudna, J. A. & Yildiz, A. A conformational checkpoint between DNA binding and cleavage by CRISPR-Cas9. Sci. Adv. 3 , eaao0027 (2017).
Gong, S., Yu, H. H., Johnson, K. A. & Taylor, D. W. DNA unwinding is the primary determinant of CRISPR-Cas9 activity. Cell Rep. 22 , 359–371 (2018).
Maji, B. et al. A high-throughput platform to identify small-molecule inhibitors of CRISPR-Cas9. Cell 177 , 1067–1079 e1019 (2019).
Nishimasu, H. et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156 , 935–949 (2014).
Jinek, M. et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 343 , 1247997 (2014).
Yamada, M. et al. Crystal structure of the minimal Cas9 from campylobacter jejuni reveals the molecular diversity in the CRISPR-Cas9 systems. Mol. Cell 65 , 1109–1121.e1103 (2017).
Zhu, X. et al. Cryo-EM structures reveal coordinated domain motions that govern DNA cleavage by Cas9. Nat. Struct. Mol. Biol. 26 , 679–685 (2019).
Das, A. et al. Coupled catalytic states and the role of metal coordination in Cas9. Nat. Catal. 6 , 969–977 (2023).
Sun, W. et al. Structures of neisseria meningitidis Cas9 complexes in catalytically poised and anti-CRISPR-inhibited states. Mol. Cell 76 , 938–952.e935 (2019).
Gasiunas, G. et al. A catalogue of biochemically diverse CRISPR-Cas9 orthologs. Nat. Commun. 11 , 5512 (2020).
Li, S. et al. Universal toxin-based selection for precise genome engineering in human cells. Nat. Commun. 12 , 497 (2021).
Dang, Y. et al. Optimizing sgRNA structure to improve CRISPR-Cas9 knockout efficiency. Genome Biol. 16 , 280 (2015).
Riesenberg, S., Helmbrecht, N., Kanis, P., Maricic, T. & Paabo, S. Improved gRNA secondary structures allow editing of target sites resistant to CRISPR-Cas9 cleavage. Nat. Commun. 13 , 489 (2022).
Chen, F. et al. Targeted activation of diverse CRISPR-Cas systems for mammalian genome editing via proximal CRISPR targeting. Nat. Commun. 8 , 14958 (2017).
Acharya, S. et al. Francisella novicida Cas9 interrogates genomic DNA with very high specificity and can be used for mammalian genome editing. Proc. Natl Acad. Sci. USA 116 , 20959–20968 (2019).
Kim, N. et al. Prediction of the sequence-specific cleavage activity of Cas9 variants. Nat. Biotechnol. 38 , 1328–1336 (2020).
Kim, H. K. et al. High-throughput analysis of the activities of xCas9, SpCas9-NG and SpCas9 at matched and mismatched target sequences in human cells. Nat. Biomed. Eng. 4 , 111–124 (2020).
Lazzarotto, C. R. et al. CHANGE-seq reveals genetic and epigenetic effects on CRISPR-Cas9 genome-wide activity. Nat. Biotechnol. 38 , 1317–1327 (2020).
Wimberger, S. et al. Simultaneous inhibition of DNA-PK and Polϴ improves integration efficiency and precision of genome editing. Nat. Commun. 14 , 4761 (2023).
Finn, J. D. et al. A single administration of CRISPR/Cas9 lipid nanoparticles achieves robust and persistent in vivo genome editing. Cell Rep. 22 , 2227–2235 (2018).
Jiang, C. et al. A non-viral CRISPR/Cas9 delivery system for therapeutically targeting HBV DNA and pcsk9 in vivo. Cell Res. 27 , 440–443 (2017).
Miller, J. B. et al. Non-viral CRISPR/Cas gene editing in vitro and in vivo enabled by synthetic nanoparticle Co-delivery of Cas9 mRNA and sgRNA. Angew. Chem. Int Ed. Engl. 56 , 1059–1063 (2017).
Lundin, A. et al. Development of an ObLiGaRe doxycycline inducible Cas9 system for pre-clinical cancer drug discovery. Nat. Commun. 11 , 4903 (2020).
Cofsky, J. C., Soczek, K. M., Knott, G. J., Nogales, E. & Doudna, J. A. CRISPR-Cas9 bends and twists DNA to read its sequence. Nat. Struct. Mol. Biol. 29 , 395–402 (2022).
Hibshman, G. N. et al. Unraveling the mechanisms of PAMless DNA interrogation by SpRY-Cas9. Nat. Commun. 15 , 3663 (2024).
Liu, M. S. et al. Engineered CRISPR/Cas9 enzymes improve discrimination by slowing DNA cleavage to allow release of off-target DNA. Nat. Commun. 11 , 3576 (2020).
Wang, Y. et al. Guide RNA engineering enables efficient CRISPR editing with a miniature Syntrophomonas palmitatica Cas12f1 nuclease. Cell Rep. 40 , 111418 (2022).
Sjogren, A. K. et al. Critical differences in toxicity mechanisms in induced pluripotent stem cell-derived hepatocytes, hepatic cell lines and primary hepatocytes. Arch. Toxicol. 88 , 1427–1437 (2014).
Clement, K. et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 37 , 224–226 (2019).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152 , 36–51 (2005).
Punjani, A. Real-time cryo-EM structure determination. Microsc. Microanal. 27 , 1156–1157 (2021).
Article ADS Google Scholar
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14 , 290–296 (2017).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60 , 2126–2132 (2004).
Croll, T. I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D. Struct. Biol. 74 , 519–530 (2018).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D. Struct. Biol. 74 , 531–544 (2018).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27 , 14–25 (2018).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30 , 70–82 (2021).
Download references
Acknowledgements
We thank Steve Rees for supporting this work. We thank the members of the Genome Engineering Department and Joanna Rejman for critically reading the manuscript and useful suggestions; Euan Gordon, Veronika Saez Jimenez, and Protein Science team for purification of recombinant PsCas9; Anders Gunnarsson for the help in establishing in vitro binding assays; George Thom and Salman Mustfa for the help with mRNA production; Kristina Friis and Kai Liu for formulating LNPs for in vivo study; Andrea Ahnmark, Annika Stenberg, Marie Johansson and Steven Oag for the help with in vivo experiments; Maryam Clausen and NGS team for supporting amplicon sequencing; Mike Firth for the help with processing NGS data; Kevin Holden for the sgRNA reagents. This work was supported by the AZ Postdoctoral Fellowship to I.W. and the National Institutes of Health grant R35GM138348 to D.W.T. and and Welch Foundation Research Grant F-1938 to D.W.T.
Author information
These authors contributed equally: Dmitrii Degtev, Jack Bravo.
Authors and Affiliations
Genome Engineering, Discovery Sciences, BioPharmaceuticals R&D Unit, AstraZeneca, Gothenburg, Sweden
Dmitrii Degtev, Aikaterini Emmanouilidi, Aleksandar Zdravković, Oi Kuan Choong, Niklas Selfjord, Isabel Weisheit, Pinar Akcakaya, Michelle Porritt, Marcello Maresca & Grzegorz Sienski
Department of Molecular Biosciences, University of Texas at Austin, Austin, TX, USA
Jack Bravo & David Taylor
Translational Genomics, Discovery Sciences, BioPharmaceuticals R&D Unit, AstraZeneca, Gothenburg, Sweden
Julia Liz Touza
Quantitative Biology, Discovery Sciences, BioPharmaceuticals R&D Unit, AstraZeneca, Gothenburg, Sweden
Margherita Francescatto
Center for Systems and Synthetic Biology, University of Texas at Austin, Austin, TX, 78712, USA
David Taylor
LIVESTRONG Cancer Institutes, Dell Medical School, Austin, TX, 78712, USA
You can also search for this author in PubMed Google Scholar
Contributions
D.D., M.M. and G.S. initiated the project. D.D. and J.B. performed most of the experimental work with the help from A.E., A.Z., O.K.C., J.L.T., N.S., I.W., M.F., P.A., M.P., M.M., D.T., G.S. provided technical input and guidance. A.E. designed and performed in vivo experimental work including animal handling, sampling and data processing. D.D. and J.B. prepared the manuscript with input from all authors. D.D., D.T., M.M. and G.S. supervised the study.
Corresponding authors
Correspondence to Dmitrii Degtev , Marcello Maresca , David Taylor or Grzegorz Sienski .
Ethics declarations
Competing interests.
D.D., A.E., A.Z., O.K.C., J.L.T., N.S., I.W., M.F., P.A., M.P., M.M. and G.S. are employees and shareholders of AstraZeneca. G.S. and M.M. are listed as co-inventors in a patent application filed by AstraZeneca Ab (application number: WO2022248645A1; status: published) related to this work covering aspects of protein engineering. J.B. and D.T. declare no competing interests. This work was supported by the National Institutes of Health grant R35GM138348 to D.T. and Welch Foundation Research Grant F-1938 to D.T; by the AstraZeneca Postdoctoral Fellowship to I.W.
Peer review
Peer review information.
Nature Communications thanks Pranam Chatterjee, Osamu Nureki, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information, peer review file, description of additional supplementary files, supplementary data 1, supplementary data 2, supplementary data 3, supplementary data 4, supplementary data 5, supplementary data 6, supplementary data 7, supplementary data 8, reporting summary, source data, source data, rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/ .
Reprints and permissions
About this article
Cite this article.
Degtev, D., Bravo, J., Emmanouilidi, A. et al. Engineered PsCas9 enables therapeutic genome editing in mouse liver with lipid nanoparticles. Nat Commun 15 , 9173 (2024). https://doi.org/10.1038/s41467-024-53418-8
Download citation
Received : 28 February 2024
Accepted : 09 October 2024
Published : 07 November 2024
DOI : https://doi.org/10.1038/s41467-024-53418-8
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.

IMAGES
VIDEO
COMMENTS
A lipid is any of various organic compounds that are insoluble in water. They include fats, waxes, oils, hormones, and certain components of and function as energy-storage molecules and chemical messengers. Together with proteins carbohydrates, lipids are one of the principal structural components of living .
Lipids are oily or greasy nonpolar molecules, stored in the adipose tissue of the body. Lipids are a heterogeneous group of compounds, mainly composed of hydrocarbon chains. Lipids are energy-rich organic molecules, which provide energy for different life processes. Lipids are a class of compounds characterised by their solubility in nonpolar ...
Classification of Lipids. Lipids can be classified according to their hydrolysis products and according to similarities in their molecular structures. Three major subclasses are recognized: 1. Simple lipids (a) Fats and oils which yield fatty acids and glycerol upon hydrolysis. (b) Waxes, which yield fatty acids and long-chain alcohols upon ...
Lesson 2: Important molecules for biology. Elements and atoms. Introduction to carbohydrates. Introduction to proteins and amino acids. Introduction to lipids. Introduction to nucleic acids and nucleotides. Introduction to vitamins and minerals. Biological macromolecules review. Biological macromolecules.
Lipid Classification For purposes of simplicity of study lipids are divided into five categories based on their function: Energy-storage lipids - A fat, triacylglycerols or triglycerides. Membrane lipids - phospholipids, sphingoglycolipids, and cholesterol Emulsification lipids - bile acids, soaps and detergents
KnowledgePath Assignment 1: Introduction to Biological Molecules. Properties of Carbon. Organic Vs Inorganic. Classification of Carbohydrates. ... Introduction to Lipids Lipids. Lipids represent a large group of molecules which consist almost entirely of carbon and hydrogen atoms. A key distinguishing feature of lipids is the fact that they are ...
What you'll learn to do: Illustrate different types of lipids and relate their structure to their role in biological systems. Fats and oils are probably the type of lipid that you're most familiar with in your everyday life. The word fat typically brings up a negative picture in our minds. In diets, we're advised to stay away from fatty ...
Lipids perform three primary biological functions within the body: they serve as structural components of cell membranes, function as energy storehouses, and function as important signaling molecules. The three main types of lipids are triglycerides, phospholipids, and sterols. Triglycerides make up more than 95 percent of lipids in the diet ...
Figure 7.1.1 7.1. 1: Fresh-caught eulachon smelt from the Kuskokwim River, Alaska, 2008. ("Kuskokwim Smelt" by Andrea Pokrzywinski is licensed under CC BY 2.0) Beyond the timing of its late winter arrival, what makes the eulachon so valuable is its high lipid content. It's so oily that dried eulachon will ignite and burn like a candle ...
In this outcome, we will discuss lipids, or fats, and the role they play in our bodies. What You'll Learn to Do. Distinguish between the different kinds of lipids; Identify several major functions of lipids; Learning Activities. The learning activities for this section include the following: Lipids; Self Check: Lipids
For example, the introduction of cardiolipin or PG could potentially improve growth, but only if these lipids are present with the correct acyl chain configurations that support optimal membrane ...
Background Colorectal cancer (CRC) is the most common gastrointestinal malignancy worldwide, with obesity-induced lipid metabolism disorders playing a crucial role in its progression. A complex connection exists between gut microbiota and the development of intestinal tumors through the microbiota metabolite pathway. Metabolic disorders frequently alter the gut microbiome, impairing immune and ...
Clinical implementation of therapeutic genome editing relies on efficient in vivo delivery and the safety of CRISPR-Cas tools. Previously, we identified PsCas9 as a Type II-B family enzyme capable ...