An official website of the United States government
Official websites use .gov A .gov website belongs to an official government organization in the United States.
Secure .gov websites use HTTPS A lock ( Lock Locked padlock icon ) or https:// means you've safely connected to the .gov website. Share sensitive information only on official, secure websites.
- Publications
- Account settings
- Advanced Search
- Journal List

DNA Extraction and Polymerase Chain Reaction
Nalini gupta.
- Author information
- Copyright and License information
Address for correspondence: Dr. Nalini Gupta, Department of Cytology and Gynaecological Pathology, Postgraduate Institute of Medical Education and Research, Chandigarh - 160 012, India. E-mail: [email protected]
This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
DNA extraction and polymerase chain reaction (PCR) are the basic techniques employed in the molecular laboratory. This short overview covers various physical and chemical methods used for DNA extraction so as to obtain a good-quality DNA in sufficient quantity. DNA can be amplified with the help of PCR. The basic principle and different variants of PCR are discussed.
Keywords: DNA extraction, Polymerase chain reaction, real time PCR
DNA E XTRACTION
DNA extraction is a method to purify DNA by using physical and/or chemical methods from a sample separating DNA from cell membranes, proteins, and other cellular components. Friedrich Miescher in 1869 did DNA isolation for the first time.
The use of DNA isolation technique should lead to efficient extraction with good quantity and quality of DNA, which is pure and is devoid of contaminants, such as RNA and proteins. Manual methods as well as commercially available kits are used for DNA extraction. Various tissues including blood, body fluids, direct Fine needle aspiration cytology (FNAC) aspirate, formalin-fixed paraffin-embedded tissues, frozen tissue section, etc., can be used for DNA extraction.
DNA extraction involves lysing the cells and solubilizing DNA, which is followed by chemical or enzymatic methods to remove macromolecules, lipids, RNA, or proteins.
DNA extraction techniques include organic extraction (phenol–chloroform method), nonorganic method (salting out and proteinase K treatment), and adsorption method (silica–gel membrane).
Organic extraction
This method is labor intensive and time consuming.
Cell lysis can be done using nonionic detergent (sodium dodecyl sulfate), Tris–Cl, and Ethylene diamine tetraacetic acid (EDTA), and this step is followed by removal of cell debris by centrifugation. Protease treatment is then used to denature proteins. Organic solvents such as chloroform, phenol, or a mixture of phenol and chloroform (phenol/chloroform/isoamyl alcohol ratio is 25:24:1) are used for denaturation and precipitation of proteins from nucleic acid solution, and denatured proteins are removed by centrifugation and wash steps. RNAse treatment is done for the removal of unwanted RNA. Precipitation with ice-cold ethanol is performed for concentrating DNA. Nucleic acid precipitate is formed, when there is moderate concentration of monovalent cations (salt). This precipitate can be recovered by centrifugation and is redissolved in TE buffer or double-distilled water.
Other methods include silica-based technology (DNA absorbs to silica beads/particles at a specific pH in presence of specific salts), magnetic separation (DNA binds reversibly to magnetic beads, which are coated with DNA-binding antibody), anion exchange technology, salting out, and cesium chloride density gradients.
Assessing the quality and yield of DNA: The quality and yield of DNA are assessed by spectrophotometry or by gel electrophoresis. Spectrophotometry involves estimation of the DNA concentration by measuring the amount of light absorbed by the sample at specific wavelengths. Absorption peak for nucleic acids is at ~260 nm. The A 260 /A 280 ratio is ~1.8 for dsDNA. A ration of less than 1.7 indicates protein contamination.
P OLYMERASE C HAIN R EACTION
Polymerase chain reaction (PCR) is a robust technique to selectively amplify a specific segment of DNA in vitro .[ 1 ] PCR is performed on thermocycler and it involves three main steps: (1) denaturation of dsDNA template at 92–95°C, (2) annealing of primers at 50–70°C, and (3) extension of dsDNA molecules at approx. 72°C. These steps are repeated for 30–40 cycles.
Various chemical components of PCR include MgCl 2 , buffer (pH: 8.3–8.8), Deoxynucleoside triphosphates (dNTPs), PCR primers, target DNA, and thermostable DNA polymerase.[ 2 ]
Target sequence is the sequence within the DNA template, which will be amplified by PCR.[ 2 ]
PCR primers are single-stranded DNA (usually 18–25 nucleotides long), which match the sequences at the ends of or within the target DNA, and these are required to start DNA synthesis in PCR.[ 2 ]
Various types of PCR
Conventional (qualitative) PCR
Multiplex PCR
Reverse transcriptase PCR and Quantitative Real-time PCR
Quantitative PCR
Hot-start PCR
Touchdown PCR
Assembly PCR
Methylation-specific PCR
LAMP assay.
Multiplex PCR: It is used to amplify multiple targets in a single PCR permitting their simultaneous analysis.
Nested PCR: It is a modified PCR intended to decrease nonspecific binding of products because of amplification of unexpected primer-binding sites. It involves two PCR steps. In the first PCR reaction, one pair of primers is used to produce DNA products, which act as a target for the second PCR reaction. It helps to increase the specificity of DNA amplification.[ 3 , 4 ]
Reverse transcriptase PCR: RT-PCR involved mRNA as the starting material and it uses reverse transcriptase to convert mRNA into the complementary DNA (cDNA). This cDNA is then amplified with the help of regular PCR.
Quantitative PCR: It is used to quantitate the amount of target DNA (or RNA) in a particular sample.
Hot-start PCR: The main advantage of hot-start PCR is to decrease nonspecific amplification of DNA at lower temperature steps of PCR. Reaction components are manually heated before adding Taq polymerase to the DNA-melting temperature (i.e. 95°C).[ 4 ]
Touchdown PCR: Annealing temperature during the first two cycles of amplification is set at approximately 3–10°C above estimated T m and the temperature is slowly decreased in the subsequent cycles. Higher annealing temperature in two initial cycles leads to more specificity for primer binding, and the lower temperatures allow more efficient amplification later on.[ 4 ]
Assembly PCR: Assembly PCR helps in synthesis of long DNA segments by doing PCR on a pool of long oligonucleotides having short overlapping segments and in turn assembling more DNA segments into one segment.
Methylation specific PCR: This PCR involves sodium bisulfite treatment and is used to identify patterns of DNA methylation at cytosine guanine islands in genomic DNA.
LAMP assay (loop-mediated isothermal amplification): It is another modification of PCR, which uses 3:6 primers sets, one of which is a loop-like primer. This technique utilizes Bst-polymerase.
Real-time PCR: It allows quantitative estimation of PCR product, as the amplification progresses. It uses nonspecific dye such as SYBR ® green I or fluorescence resonance energy transfer.
PCR products are then sequenced to determine the order of bases in the DNA segment.
Financial support and sponsorship
Conflicts of interest.
There are no conflicts of interest.
R EFERENCES
- 1. Lo AC, Feldman SR. Polymerase chain reaction: Basic concepts and clinical applications in dermatology. J Am Acad Dermatol. 1994;30:250–60. doi: 10.1016/s0190-9622(94)70025-7. [ DOI ] [ PubMed ] [ Google Scholar ]
- 2. Clark DP, Pazdernik NJ. Molecular Biology, Polymerase Chain Reaction. 2nd ed. United States of America (USA): Elsevier BV; 2013. pp. 163–93. Chap. 6. [ Google Scholar ]
- 3. Niemz A, Ferguson TM, Boyle DS. Point-of-care nucleic acid testing for infectious diseases. Trends Biotech. 2011;29:240–50. doi: 10.1016/j.tibtech.2011.01.007. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 4. Lorenz TC. Polymerase chain reaction: Basic protocol plus troubleshooting and optimizing strategies. J Vis Exp. 2012;63:e3998. doi: 10.3791/3998. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- View on publisher site
- Collections
Similar articles
Cited by other articles, links to ncbi databases.
- Download .nbib .nbib
- Format: AMA APA MLA NLM
Add to Collections

Nucleus of Notes, News, and Update

DNA extraction- Introduction, Methods, History, Types, Confirmation, Detection
Table of Contents
Introduction:
- Since its beginnings in 1869, Deoxyribonucleic Acid (DNA) extraction has progressed significantly. Many of the potential downstream applications in the field of molecular biology require it as the first step. (PCR, DNA cloning, DNA sequencing, DNA electrophoresis) Next-generation sequencing, PCR, DNA cloning, DNA sequencing, DNA electrophoresis)
- DNA extraction is a technique for isolating DNA from cell membranes, proteins, and other biological components from a sample using physical and/or chemical processes. Several parameters, such as tissue type and DNA integrity, must be considered when selecting a DNA extraction method.
- Phenol-chloroform isoamyl alcohol, Proteinase K, CTAB method, spin column-based approaches, and magnetic bead-based technology are some of the different types of DNA extraction methods available. The procedure to utilize, on the other hand, is determined by the sample type and the purity and quantity of DNA we wish to obtain.
- Every DNA extraction process differs depending on the type of sample; for instance, plant DNA extraction differs from blood DNA extraction. Similarly, the process for isolating bacterial DNA differs from those used for other kinds. As a result, different DNA isolation procedures are required for various samples.
In 1869, Friedrich Miescher made the first attempt at DNA extraction. He extracted the cell substance and termed it “nuclei,” which was later named “nucleic acid” by his trainee. He invented a method for nucleic acid isolation by accident, but he wasn’t sure if the nucleic acid he extracted was DNA.
Meselson and Stahl later established a full-function DNA extraction procedure in 1958. The first methodology for recovering DNA from E. coli bacteria was the density gradient centrifugation protocol.
Lahiri and Nurnberger introduced the proteinase K enzyme technique of DNA extraction protocol in 1991. They even used the Nonidet P40 and SDS to modify the technique. Miller et al., however, reported on the use of proteinase K in DNA extraction in 1988.
Joseph Sambrook and David W. Russell pioneered the phenol-chloroform isoamyl alcohol technique, which has become increasingly popular in recent years. Among researchers, the current method is the most popular .
What is DNA extraction?
“Extracting DNA from cells” is the most basic definition of DNA extraction. Depending on the method, chemical, and test employed, we can explain it in a variety of ways. Here are a few different definitions of DNA extraction:
“A DNA extraction is the process of isolating DNA by breaking the cell wall/cell membrane and the nuclear membrane.”
“A DNA extraction is described as the separation of DNA from the cell membrane and nuclear membrane using chemicals, enzymes, or physical disturbances.”
“Nucleic acid extraction” or “DNA extraction” is the process of extracting nucleic acid from the rest of the cell organelle.
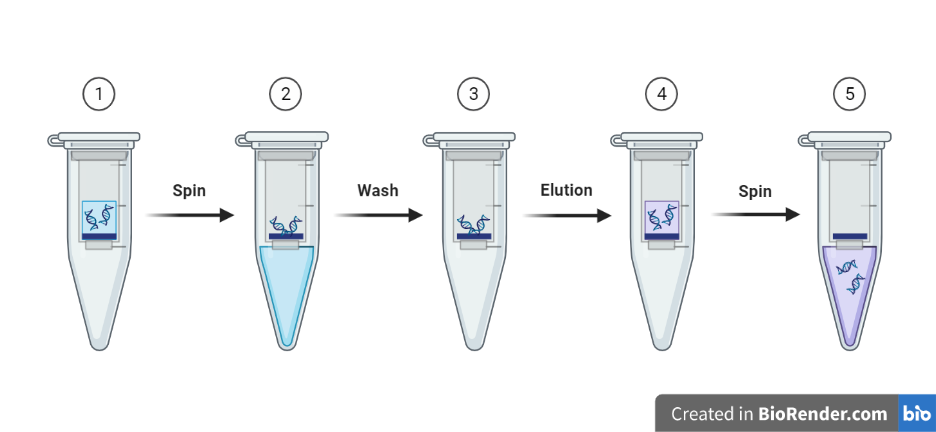
Fig: Outline of DNA extraction
How to obtain DNA?
The cytoplasm and cell membrane/cell wall make up the cell. Several organelles, such as mitochondria, ribosomes, nucleus, and endoplasmic reticulum, are found in the cytoplasm. The cell wall is absent in animal cells, although it is present in plant cells and (most) bacterial cells.
To isolate DNA-deoxyribonucleic acid, we must first breach the cell wall/cell membrane, as well as the nuclear envelope. Other cellular organelle debris must also be removed. Precipitation and purification of the DNA are the final steps.
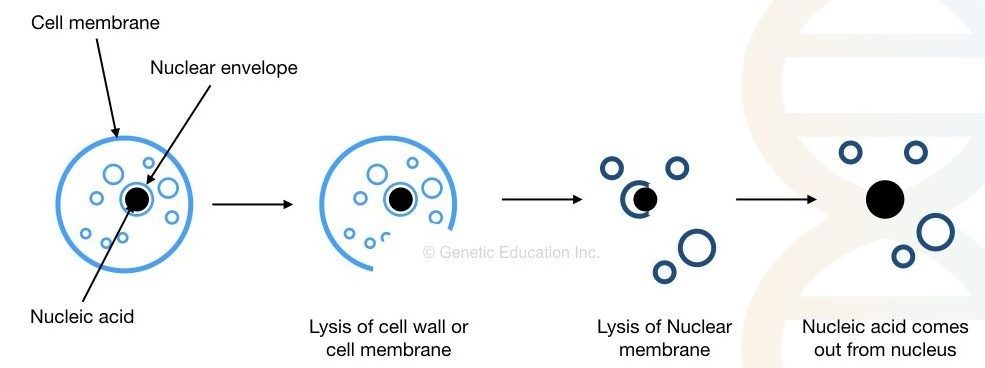
Fig : Extraction of DNA
In other words, cell lysis, precipitation, and dissolving DNA are the three major processes in the DNA isolation process. The following are the outline of the process.
Lysis of cell wall/ cell membrane
- Chemical disruption
- enzymatic disruption
- Mechanical disruption
Lysis of nuclear membrane:
- Chemical lysis
- Enzymatic lysis
Removing cell debris
- Centrifugation
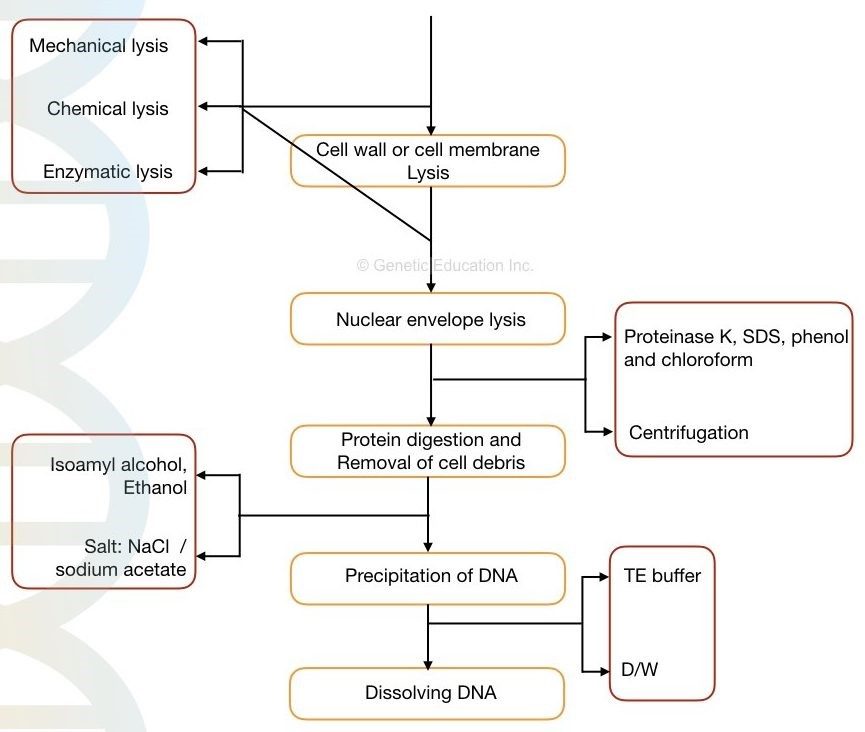
Fig : Different principle of DNA extraction
For various objectives, different compounds and combinations of chemicals are used. Protein and lipids almost entirely make up the nuclear and cell membranes. As a result, the same compounds can be used for both.
By solubilizing the cell wall/cell membrane, chemicals like SDS, CTAB, Tris, and other detergents can lyse them. Every chemical serves a certain purpose.
Proteinase K, peptidase, and protease are enzymes that break down proteins. Because it directly targets amino acid linkages and digests the protein, the enzyme functions better than any other chemical.
Furthermore, the procedure for extracting DNA varies based on the cell type.
Cells with a soft cell wall:
Some bacteria have a cell wall that is exceedingly smooth and fragile. M. tuberculosis , for example, has a smooth cell wall. We can lyse the cell wall by gently heating the bacterial solution.
The supernatant can be used directly for PCR. We can also achieve a good quality PCR result by directly placing the bacterial culture into the PCR tube for 15 minutes at first denaturation.
The use of a simple lysis buffer during the heating process, on the other hand, will enhance the yield and quality of DNA.
Cells with a harder cell wall:
The cell wall of plant cells contains pectin and other polysaccharides. The cell is protected against mechanical harm by pectin. As a result, pectin gives the plant cell wall added strength.
Hard cell walls are also found in some fungi, algae, and bacteria, which help them survive in tough environments. We must adjust the process using a combination of mechanical, chemical, and enzymatic methods in order to extract DNA from this type of cell.
In order to isolate DNA from pectin-rich cells, mechanical lysis is necessary. Mortar, pestle, grinding, and liquid nitrogen are all employed to accomplish this.
Different types of DNA extraction methods :
DNA extraction methods are broadly categorized into two categories:
- Chemical-based DNA extraction method.
- Silica-based DNA extraction method.
The chemical DNA extraction methods are also known as solution-based methods whilst solid-phase DNA extraction is a type of physical method.
Chemical or solution-based DNA extraction method:
Various organic and inorganic solutions are used in a chemical or solution-based technique. The main steps in all procedures, namely cell lysis, precipitation, and elution, are the same.
Several popular chemicals used in the solution-based DNA extraction approach include SDS, CTAB, phenol, chloroform, isoamyl alcohol, Triton X100, guanidium thiocyanate, Tris, and EDTA.
Organic solvent-based DNA extraction and inorganic solvent-based DNA extraction are two types of solution-based (also known as chemical) DNA extraction.
Organic solvents such as phenol and chloroform are used in the organic solvent-based DNA extraction method. The current procedure is not recommended due to the dangers of phenol and chloroform. Regardless, the phenol-chloroform technique is the most effective.
The proteinase-K DNA extraction process yields a higher quantity of DNA; however, it takes a long time. Proteinase-K cannot be used for a longer period of time if it is not kept cold in a cold chain. Another important drawback with this procedure is the enzyme’s decreased stability.
The proteinase K-based approach for DNA isolation is an inorganic solution-based technology.
The salting-out DNA extraction method:
Salts such as sodium chloride, potassium acetate, and ammonium acetate are used in the salting-out DNA extraction process. However, when used in conjunction with proteinase K, the procedure produces outstanding results.
The purity of the salting-out process is one of its key drawbacks; while a sufficient yield can be reached, the quality gained may not be satisfactory.
Phenol-chloroform method of DNA extraction:
This is one of the most effective ways for extracting DNA. If we perform the PCI procedure properly, the yield and purity of DNA obtained are excellent. The PCI method of DNA extraction is also known as the phenol-chloroform-isoamyl alcohol method.
Liquefaction buffer, phenol, and chloroform are the most common chemicals used in PCI DNA extraction procedures.
Tris, EDTA, MgCl2, NaCl, SDS, and other salts are present in the lysis buffer. The components of lysis buffer aid in the lysis of both the cell membrane and the nuclear envelope in this situation. The technique’s chemical components, phenol and chloroform, denature the protein portion of cells.
Role of chemicals:
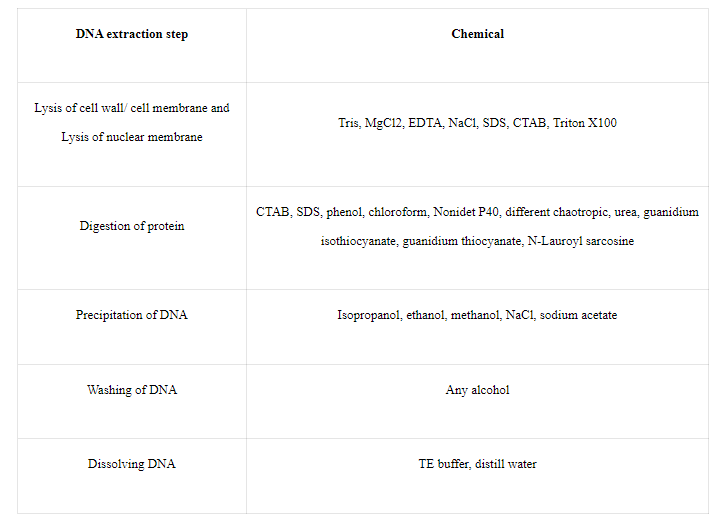
Fig: Role of several chemicals used in DNA extraction
Enzymatic method of DNA extraction:
This method is actually a combination of a salt method and an enzymatic method. Before proceeding with the enzymatic digestion, the extraction buffer is used.
The extraction buffer’s composition varies each lab, but the main ingredients are Tris, EDTA, NaCl, sodium lauryl sulfate, and SDS. No phenol, chloroform, or isoamyl alcohol is utilized in this experiment. Instead, the material is digested with the enzyme proteinase K.
Proteinase K is added to the sample and incubated for two hours, digesting all of the protein present.
The sample is immediately precipitated by cold alcohol after digestion with proteinase K. All other cell debris is removed from the sample by centrifugation. The DNA pellet is finally dissolved in TE buffer.
This DNA extraction procedure is quick and simple. We can utilize a DNA extraction buffer that is ready to use. The yield is also very high. The purity of DNA, on the other hand, is a serious concern for this approach.
Silica column-based DNA extraction method:
The silica column-based DNA extraction method is distinct from other DNA extraction techniques.
The proteinase K method requires centrifuging the sample several times and collecting the aqueous phase or pellets, depending on the extraction stage. We may need to collect an aqueous phase or pellets on occasion.
The silica-based DNA extraction method relies on the unique chemistry of silica and DNA interaction. During centrifugation, positively charged silica particles bind to negatively charged DNA and retain it in position.
The method was first described by McCormick in 1989. However, the idea was developed in 1979, when silica was used in DNA purification by Vogelstein.
The commercially accessible silica-based solid-phase DNA extraction technology is now widely employed in diagnostic laboratories. It is widely accepted because to its high-quality DNA yield and simple operating system.

Fig: Silica column-based DNA extraction method:
Application :
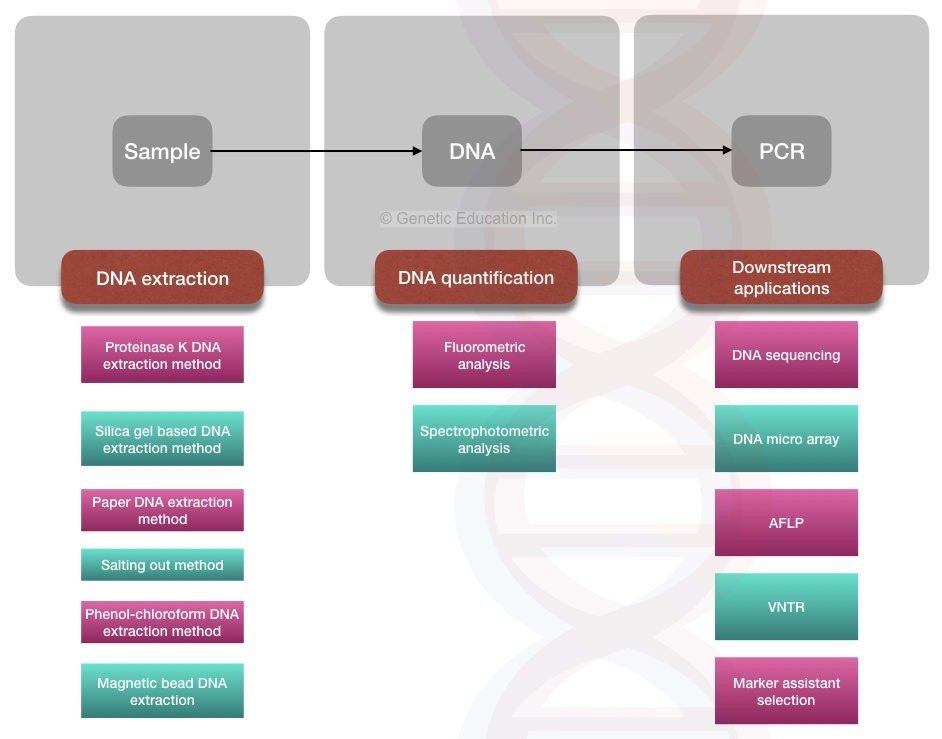
Fig: Application of DNA extraction
Method for conformation of DNA extraction/Quantification of extracted DNA:
DNA can be validated by electrophoresing on an agarose gel using ethidium bromide, or another fluorescent dye that reacts with DNA, and examining under UV light.
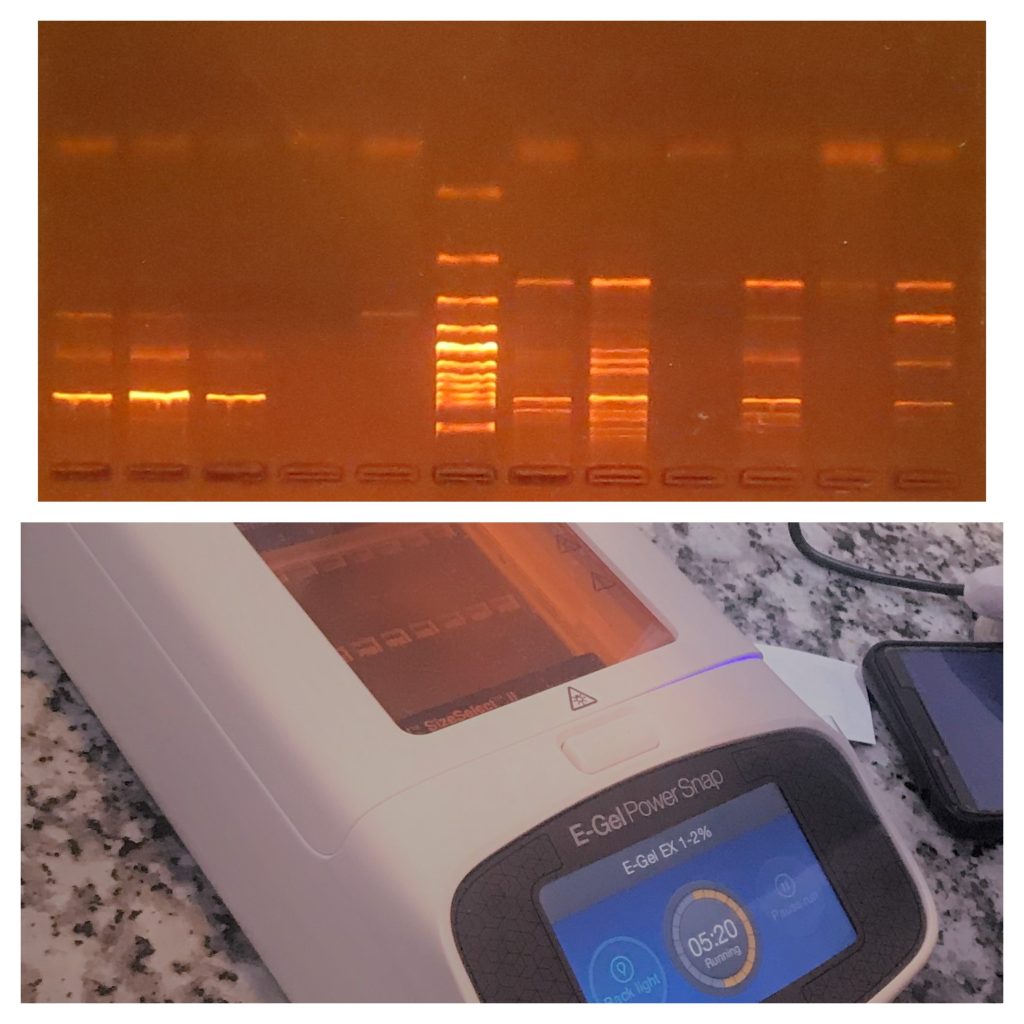
Fig: Agarose gel elctrophoresis and E-Gel electrophoresis
Absorbance Methods
- Measuring the intensity of absorbance of the DNA solution at wavelengths 260 nm and 280 nm is used as a measure of DNA purity.
- The most common purity calculation is the ratio of the absorbance at 260nm divided by the reading at 280nm. Good-quality DNA will have an A260/A280 ratio of 1.7–2.0.
- A reading of 1.6 does not render the DNA unsuitable for any application, but lower ratios indicate more contaminants are present.
Fluorescence Methods
Fluorescence measurement is another popular method for determining DNA yield and concentration due to the widespread availability of fluorometers and fluorescent DNA-binding dyes. Fluorescence methods are more sensitive than absorbance methods, especially for low-concentration samples, and DNA-binding dyes allow for more precise DNA quantification than spectrophotometric approaches.
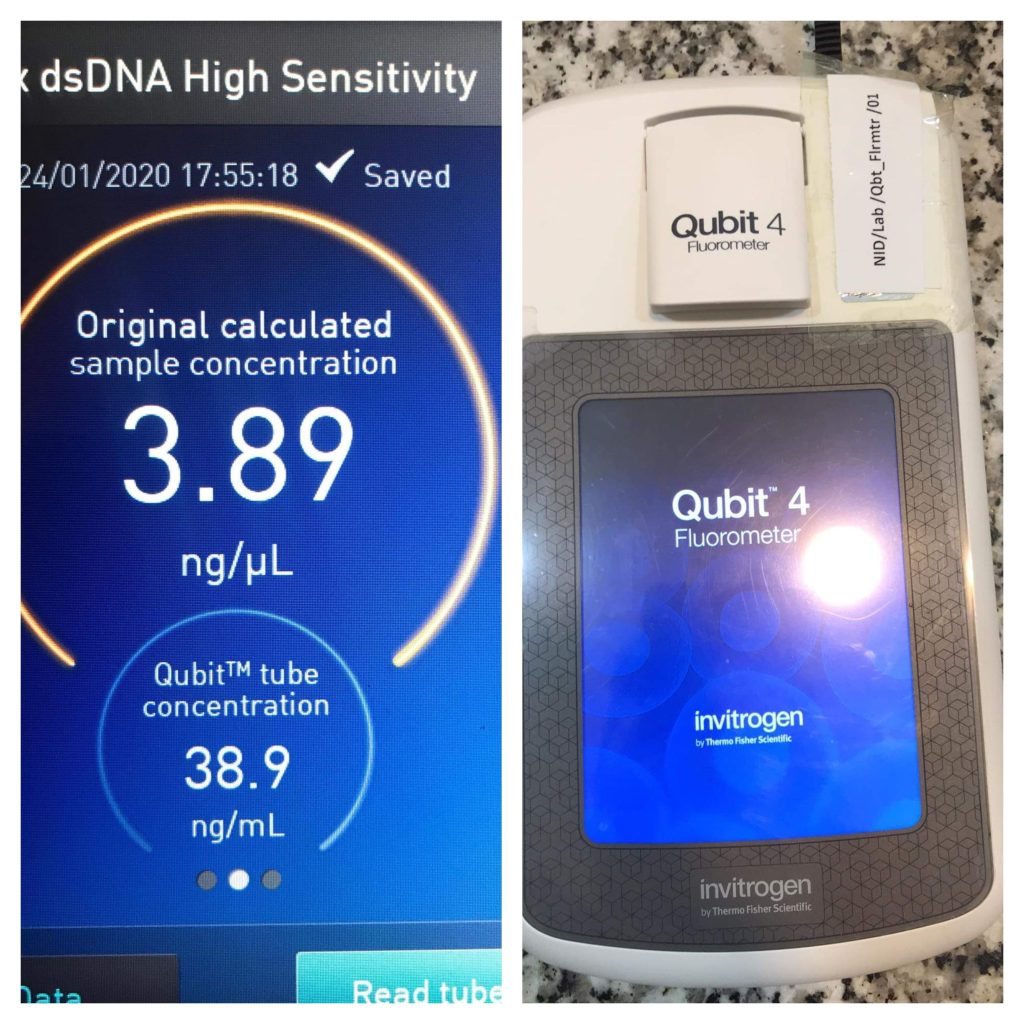
Fig: Fluorescence method for determining DNA yields
Detection of DNA:
- A diphenylamine (DPA) indicator will confirm the presence of DNA.
- This procedure involves chemical hydrolysis of DNA: when heated (e.g. ≥95 °C) in acid, the reaction requires a deoxyribose sugar and therefore is specific for DNA.
- Under these conditions, the 2-deoxyribose is converted to w-hydroxylevulinyl aldehyde, which reacts with the compound, diphenylamine, to produce a blue coloured compound.
- DNA concentration can be determined measuring the intensity of absorbance of the solution at the 600 nm with a spectrophotometer and comparing to a standard curve of known DNA concentrations.

References:
- Niemz A, Ferguson TM, Boyle DS. Point-of-care nucleic acid testing for infectious diseases. Trends Biotech. 2011; 29:240–50.
- Carpi FM, Di Pietro F, Vincenzetti S, Mignini F, Napolioni V. Human DNA extraction methods: patents and applications. Recent Pat DNA Gene Seq . 2011;5(1):1–7.
- Dahm R. Friedrich Miescher and the discovery of DNA. Dev Biol . 2005;278(2):274–288.
- Sheila Spada, … Erik Wennerberg, in Methods in Enzymology, 2020
- Dieki, R., Nsi Emvo, E. and Akue, J.P., 2022. Comparison of six methods for Loa loa genomic DNA extraction. PloS one, 17(3), p.e0265582.
- Kumar, M. and Mugunthan, M., 2018. Evaluation of three DNA extraction methods from fungal cultures. medical journal armed forces india, 74(4), pp.333-336.
- Abdel-Latif, A. and Osman, G., 2017. Comparison of three genomic DNA extraction methods to obtain high DNA quality from maize. Plant Methods, 13(1), pp.1-9.
Leave a Comment Cancel Reply
Your email address will not be published. Required fields are marked *
Save my name, email, and website in this browser for the next time I comment.
biotechnology?
DNA extraction
DNA extraction is the technique used to isolate DNA in a biological sample.
As shown in this photo, DNA, a long stringy molecule, can be lifted out of a solution by the use of a glass rod or wooden stick which it naturally wraps around when turned.

The extraction of DNA is pivotal to biotechnology. It is the starting point for numerous applications, ranging from fundamental research to routine diagnostic and therapeutic decision-making. Extraction and purification are also essential to determining the unique characteristics of DNA, including its size, shape and function.
DNA was first isolated by the Swiss physician, Friedrich Miescher, in 1869 while working in the laboratory of the biochemist Felix Hoppe-Seyler. This he did as part of a project to determine the chemical composition of cells which he saw as the means to unravelling the fundamental principles of the life of cells. Initially, he began this research by using lymphocytes drawn from lymph nodes but was unable to get sufficient quantities for analysis so switched to using leucocytes, white blood cells, which he gathered from pus found on fresh surgical bandages collected from a nearby surgical clinic. In the course of his work on leucocytes he noticed the precipitation of a substance when acid was added and that this dissolved following the addition of alkali. Mierscher decided to call the new substance 'nuclein' by virtue of its presence in the nuclei of the cell. Upon further analysis, Miescher noted that the chemical composition of the substance differed from proteins and other known molecules. He speculated that it played a central role in cells and was involved in the division of cells. Following this, Miescher developed a method for isolating nuclein from salmon sperm. Many advances have been made to the methods for extracting and purifying DNA since Miescher's time. From the 1950s routine laboratory extraction of DNA became reliant on the use of density gradient centrifuges. Until very recently most methods for extracting DNA remained complex, labour-intensive and time-consuming. They also provided only small quantities of DNA. Today there are many specialised extraction methods. These are generally either solution-based or column-based. The extraction of DNA has become much easier with the emergence of commercial kits and the automation of the process. Such changes have both sped up production and increased the yield of DNA.

Application
The ability to extract DNA is of primary importance to studying the genetic causes of disease and for the development of diagnostics and drugs. It is also essential for carrying out forensic science, sequencing genomes, detecting bacteria and viruses in the environment and for determining paternity.
Science links: Science home | Cancer immunotherapy | CRISPR-Cas9 | DNA | DNA polymerase | DNA Sequencing | Epigenetics | Faecal microbiota transplant | Gene therapy | Immune checkpoint inhibitors | Infectious diseases | Messenger RNA (mRNA) | Monoclonal antibodies | Nanopore sequencing | Organ-on-a-chip | p53 Gene | PCR | Phage display | Phage therapy | Plasmid | Recombinant DNA | Restriction enzymes | Stem cells | The human microbiome | Transgenic animals |
Respond to or comment on this page on our feeds on Facebook , Instagram , Mastodon or Twitter .
- About What is Biotechnology
- Advisory Board
- © 2024

- ISI Impact Factor: 1.042
- [email protected]
- Volume 8 - Issue 1
- Download PDF [ P: 39-45 ]
Article Citation
Preetha J Shetty, The Evolution of DNA Extraction Methods. 2020 - 8(1). AJBSR.MS.ID.001234. DOI: 10.34297/AJBSR.2020.08.001234.
Navigation Menu
Introduction, materials and methods, discussion commentary, acknowledgements, conflict of interest, share this article.

The Evolution of DNA Extraction Methods
Mariyam dairawan, preetha j shetty*.
*Corresponding author: Dr. Preetha J Shetty, Associate Professor, Department of Biomedical Sciences, College of Medicine, Gulf Medical University, UAE.
Received: February 18, 2020; Published: March 11, 2020
DOI: 10.34297/AJBSR.2020.08.001234
Since the first DNA extraction performed by Friedrich Miescher in 1869, scientists have made extraordinary progress in designing extraction methods that are more reliable, easier and faster to perform, more cost-effective and produce a higher yield. The classic liquid-liquid DNA extraction method involves the use of organic and inorganic reagents such as phenol-chloroform which pose a toxic threat to humans. Many newer techniques are now based on physical extraction, which has significantly contributed to developing simpler methods for DNA handling, such as extraction using magnetic beads and cellulose-based filter paper. With the advent of gene-editing and personalized medicine, there has been an increase in the demand for reliable and efficient DNA isolation methods that can yield adequate quantities of high-quality DNA with minimal impurities. The current review addresses the evolution of different DNA extraction techniques from solvent-based methods to physical extraction methods each with its varying set of advantages and limitations.
Keywords: DNA; DNA Extraction Methods; DNA Isolation; Purification
Abbreviations: SEC: Size Exclusion Chromatography; IEC: Ion Exchange Chromatography; AC: Affinity Chromatography; DNA: Deoxyribonucleic Acid; RNA: Ribonucleic Acid; SDS: Sodium Dodecyl Sulphate; EDTA: Ethylenediaminetetraacetic Acid
Extraction of nucleic acids is the starting point in any molecular biology study and hence is considered as a crucial process. The first crude extraction of DNA had been performed by the Swiss physician Friedrich Miescher in 1869. He had accidentally purified DNA from the nucleus while investigating proteins from leukocytes and found that the property of this substance was fundamentally different than proteins, hence coined the term “nuclein” [ 1 ]. DNA can be extracted from sources as diverse as clinical samples such as fine needle aspirates of body fluids and biopsy samples; forensic samples such as dried blood spots, buccal swabs and fingerprints; to soil, plant and animal tissue, insects, protozoa, bacteria and yeast [ 2 ].
Biological information is stored in DNA through its linear sequence of polynucleotides. It is translated into mRNA and subsequently transcribed into a sequence of amino acids that determines the three-dimensional structure of the protein, which in turn determines its biological function. Hence, DNA provides the instructions to maintain biological function and is considered to be the blueprint of life [ 3 ].
At neutral pH and physiological salt concentration, DNA mostly exists in the B-form, the classic right-handed double helical structure. A-form nucleic acid, found in RNA-DNA and RNA-RNA duplexes, is thicker with its base pairs packed closer together due to the syn conformation of its deoxyribose sugar ring [ 4 ]. Z-DNA is present in small amounts in the cell and has a left-handed helical structure that is thinner with more base pairs per turn as compared to B-form. Its phosphodiester backbone follows a zigzag pattern owing to the alternate stacking of purine bases that maintain a syn conformation and pyrimidine bases that are rotated into the anticonformation [ 5 ].
While double-stranded DNA is present in almost all organisms, single-stranded DNA is found in very few patho-genic viruses, such as members of the families Parvoviridae, Circoviridae, Anelloviridae and Geminiviridae [ 6 ] but is also present in bacteria and cells of While double-stranded DNA is present in almost all organisms, single-stranded DNA is found in very few patho-genic viruses, such as members of the families Parvoviridae, Circoviridae, Anelloviridae and Geminiviridae [ 6 ] but is also present in bacteria and cells of
DNA isolated from various biological samples can be used for a vast array of downstream applications, namely DNA sequencing, polymerase chain reaction (PCR), quantitative PCR (qPCR), southern blotting, random amplification of polymorphic DNA (RAPD), preparation for genomic libraries as well as amplified fragment length polymorphism (AFLP), restriction fragment length polymorphism (RFLP), short tandem repeat polymorphism (STRP), single nucleotide polymorphism (SNP) and variable number of tandem repeat (VNTR) applications [ 2 ].
In the field of medicine, these downstream applications can serve as a basis for diagnosing genetic diseases or identifying carrier status, gene therapy, pharmacogenomics to predict drug efficacy or adverse drug reactions [ 8 ] using recombinant DNA technology to derive pharmaceutical products such as hormones and vaccines [ 9 ] as well as predictive genetic testing for risk assessment and the use of targeted screening and early prevention strategies. For example, early detection of mutations in the RET proto-oncogene and pre symptomatic intervention is used to prevent the development of multiple endocrine neoplasia type 2 [ 10 ].
DNA fingerprinting is widely utilized in forensic science for identity verification of criminal suspects, paternity tests and in determining the lineage. In microbiology, nucleic acid analysis can be used to study phylogenetic relationships and help identify mutations that give rise to antibiotic resistance[ 10 ]. There are numerous other possible applications for isolated nucleic acid, but an innovation that has revolutionized research in genetics is the emergence of precise gene-editing technology, which has made significant contributions to the field of medicine, agriculture and synthetic biology [ 11 ].
The principal behind DNA extraction consists of the following steps: (1) disruption of cytoplasmic and nuclear membranes; (2) separation and purification of DNA from other components of the cell lysate such as lipids, proteins, and other nucleic acids; and (3) concentration and purification of DNA. When choosing a suitable method for DNA extraction, it is crucial to ensure the quality and quantity of the isolated DNA to carry out the intended downstream applications. Other factors that should be taken into consideration to optimize the DNA extraction method include the time, cost, potential toxicities, yield, laboratory equipment and expertise requirements, as well as the required sample amount for the protocol [ 13 ]. Over a period of time different isolation methods have evolved ( Figure 1 ) and the current review discusses the various isolation methods each with its set of advantages and disadvantages.
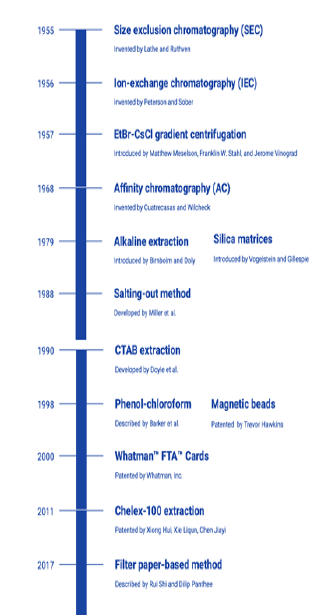
Figure 1: Timeline of the development of different DNA extraction techniques and their inventors.
Different DNA extraction methods were searched for in electronic databases, hand searched journals and through contact with authors directly. The screening and data extraction were conducted by two reviewers independently. Electronic data sources used include PubMed, ScienceDirect, ResearchGate and Google Scholar. Main search terms used were “DNA extraction methods”, “advantages of DNA extraction methods” and “limitations of DNA extraction methods”, “DNA isolation methods”, “DNA purification methods”, “solvent-based DNA extraction” and “physical DNA extraction”. Review of literature published from 1955 to till date was conducted, with a total of 116 publications reviewed.
Chromatography-Based DNA Extraction Method
Chromatography-based DNA extraction methods can be used to isolate DNA from any kind of biological material [ 14 ]. This method includes size exclusion chromatography (SEC), invented by Lathe and Ruthven (1955), ion-exchange chromatography (IEC) developed by Peterson and Sober (1956) and affinity chromatography (AC) reported by Cuatrecasas & Wilcheck [ 15 , 16 , 17 ]. In size exclusion chromatography (SEC), molecules are separated according to their molecular sizes and shape. The term gel-filtration chromatography is used when an aqueous solution is utilized to transport the DNA-containing sample through the chromatography column, as compared to gel permeation chromatography, in which an organic solvent is used instead. The column contains porous beads composed of polyacrylamide, dextran or agarose. When the sample is applied on the top and passed through the column, smaller molecules such as mRNA and proteins enter through the small pores and channels of the beads, while DNA is excluded from entering the beads and evades the matrix with its larger hydrodynamic volume. Consequently, DNA is eluted from the column faster than the smaller molecules [ 18 , 19 ]. SEC is suitable for use on substances that are sensitive to alterations in pH and metal ion concentrations [ 20 ].
Another DNA extraction method based on chromatography is ion-exchange chromatography (IEC). The column is first equilibrated with a solution containing DNA anion-exchange resin, which is used to selectively bind DNA with its positively charged diethylaminoethyl cellulose (DEAE) group. DNA is retained in the column while other cellular components such as proteins, lipids, carbohydrates, metabolites and RNA are eluted with medium-salt buffers. DNA can then be recovered by decreasing the pH or using high-salt buffers [ 20 ]. This method is relatively simple to perform when compared to other extraction methods yielding high-quality DNA, such as CsCl-gradient centrifugation [ 14 ].
Nucleic acid purification can also be achieved by affinity chromatography (AC), consisting of a similar protocol to IEC, but instead involves the use of oligo(dT) or other substances that form highly specific interactions with nucleic acid to separate it from the cell lysate. However, this protocol is mainly used for the isolation of mRNA. It is time-efficient and yields a good quantity of nucleic acid.
EtBr-CsCl Gradient Centrifugation Method
This method was developed back in 1957 by Matthew Meselson, Franklin W. Stahl, and Jerome Vinograd [ 22 ]. DNA is first mixed with cesium chloride (CsCl), the solution is then ultra-centrifuged at high speed (10,000 to 12,000 rpm) for more than 10 hours. With centrifugation, DNA separates from the rest of the substances based on its density. Depending on DNA types varying by density, one or more DNA bands appear upon reaching the isopycnic point. Ethidium bromide (EtBr) acts as an intercalating agent and is incorporated comparatively more into non-supercoiled than supercoiled DNA molecules, hence allowing supercoiled DNA to accumulate at lower densities. Location of the DNA can be easily visualized under ultraviolet light. EtBr and CsCl are removed prior to precipitation of DNA with ethanol.
This method can be used to extract DNA from bacteria, but a large amount of the material source is required. Moreover, this method is complicated, time consuming and costly due to the long duration of high-speed ultra-centrifugation required [ 23 ].
Alkaline Extraction
First introduced in 1979 by Birnboim and Doly, the method is primarily used to extract plasmid DNA from bacterial cells [ 24 ]. The sample is first suspended in an alkaline solution containing NaOH and SDS detergent for cell membrane lysis and protein denaturation. The alkaline condition selectively denatures chromosomal DNA which has a higher molecular weight than the intact plasmid DNA that remains double-stranded. Potassium acetate is then added to neutralize the solution, causing the chromosomal DNA to renature and precipitate from the solution. Plasmid DNA in the supernatant can be recovered after centrifugation [ 25 ]. A limitation to this method is the susceptibility for the plasmid DNA to be contaminated with fragmented chromosomal DNA.
Silica Matrices
high affinity between silicates and DNA was first described by Vogelstein and Gillespie in 1979 [ 26 ]. This technique is based on the principle of selective binding of the negatively charged DNA with the silica surface that is covered with positively charged ions. With the DNA tightly bound to the silica matrix, the rest of the cellular contaminants can be washed out before the extracted DNA is eluted from the silica particles using distilled water or a buffer such as Tris-EDTA [ 27 ]. Various modified protocols of this technique have been described in literature, such as the use of hydrated silica matrix for DNA extraction [ 28 ].
The silica matrices method is simple, fast to perform, cost-efficient, produces high quality DNA and can be employed for automation. Commercially available kits that involve the use of silica matrices include Thermo Fisher Purelink Genomic DNA extraction kit and QIAGEN DNeasy Blood [ 2 ]. The major limitation is that the silica matrices cannot be reused as with the resins used in the anion exchange method due to their reduced binding capacity and attachment to DNA particles even after elution. However, newer methods such as maxXbond have been designed to allow multiple use of the silica matrices [ 29 ].
Cetyltrimethylammonium Bromide (CTAB) Extraction
The CTAB extraction method was introduced by Doyle et al. in 1990 [ 32 ]. In this method, DNA-containing samples are added to 2% CTAB at alkaline pH. In a solution of low ionic strength, the extraction buffer precipitates DNA and acidic polysaccharides from the rest of the cellular components. Solutions with high salt concentrations are then used to remove DNA from the acidic polysaccharides which form a precipitate with CTAB. Hence, this method is particularly useful for DNA extraction from plants and bacteria that produce high amounts of polysaccharides [ 20 ]. DNA is then purified using various organic solvents and alcohols, including toxic agents such as phenol and chloroform. Major limitations to this method include the use of toxic reagents and its time-consuming protocol [ 14 ].
Phenol-chloroform method
The phenol-chloroform DNA extraction method was introduced in 1998 by Barker et al. [ 37 ]. Cells are first treated with a lysis buffer containing detergents such as sodium dodecyl sulphate (SDS) to dissolve cell membranes and the nuclear envelope. Other components of the lysis buffer can include 10 mM Tris, 1 mM EDTA and 0.1 M NaCl. Phenol-chloroform-isoamyl alcohol (PCIA) reagent is then added in a ratio of 25:24:1 [ 16 ]. Both SDS and phenol denatures the proteins efficiently, and isoamyl alcohol prevents emulsification and hence facilitates precipitation of DNA.
The contents are mixed to form a biphasic emulsion followed by vortexing if the DNA molecules to be extracted are <10 kb or by gentle shaking if DNA molecules are 10–30 kb. The emulsion separates into two phases upon centrifugation: the upper aqueous phase composed of DNA dissolved in water and the bottom organic phase containing organic solvents and hydrophobic cellular components including proteins. The aqueous portion is then transferred to a fresh tube with the use of a pipette, and the organic phase can be discarded. Aspiration of organic solvents can be avoided by leaving behind a layer of the aqueous phase close to the interface. These steps are repeated until the interface between the aqueous and organic phases is cleared from protein [33].
Chloroform increases the density of the organic phase, preventing phenol from inverting into the aqueous phase, which might occur without the use of chloroform due to its close density to water (1 g/mL). Hence, chloroform is used to preserve DNA from being degraded by phenol [ 34 ].
DNA can be concentrated via the standard ethanol precipitation method by adding sodium acetate solution (0.3M final concentration, pH 5.2) and ethanol in 2:1 or 1:1 ratio, followed by centrifugation to separate DNA from the solution. The pellet is washed with cold 70% ethanol to eliminate excess salt from the DNA. Finally, the DNA pellet is centrifuged again before the removal of ethanol. The pellet is dried and resuspended either in sterile distilled water or aqueous buffer [16].
The phenol-chloroform method is the gold-standard for DNA extraction. It can be used to extract DNA from blood, suspension culture, or tissue homogenate. It provides a high yield and is relatively inexpensive with a cost estimate of less than $5 for one sample.16 However, the toxic nature of phenol and chloroform necessitates the use of fume hoods and is a major limitation to this method. Extracted DNA samples are higher in purity than other conventional methods but lower than those obtained using columnbased methods [ 10 ]. An example of a commercially available kit based on this method is Thermo Fisher Easy-DNA® [ 2 ].
Magnetic beads
In 1998, Trevor Hawkins filed a patent titled “DNA purification and isolation using magnetic particles” [ 37 ]. Magnetic nanoparticles coated with a DNA binding antibody or polymer with specific affinity to DNA can be employed to bind DNA to its surface. Magnetic beads are generally composed of magnetite or maghemite at the core, and surface substances that can be used include silica as well as functional groups such as sulphate and hydroxyl groups [ 38 ]. Separation of the DNA-bound magnetic beads from the cell lysate can be achieved by applying a magnetic field at the bottom of the tube using an external magnet. With the beads aggregated at the bottom of the tube, the supernatant can then be rinsed away. The magnetic pellet can be eluted via the ethanol precipitation method, after which the pellet is incubated at 65°C to elute the magnetic particles from the DNA [ 16 ]. The DNA yield obtained by this method is comparable to that obtained in the other conventional methods and the protocol has been shown to take less than 15 minutes to complete, which is much faster as compared to other conventional methods that can take up to several hours [ 39 ]. Furthermore, it is ideal for automation and require little equipment to perform as it does not depend on centrifugation. Another advantage over centrifugation-based methods is that this technique does not involve the use of shear forces that could damage the integrity of nucleic acids. However, this protocol is not cost effective as compared to other methods. [ 14 , 40 ]
Cellulose-based paper
Cellulose is an extensively hydroxylated polymer that has a high binding affinity for DNA. In 2000, Whatman, Inc. filed a patent titled “Ftacoated media for use as a molecular diagnostic tool” [41]. The Whatman™ FTA™ Cards are an example of commercially available cellulose-based paper that is widely used for DNA extraction. They are impregnated with buffers, detergents, and chelating agents that facilitate the extraction of DNA [ 42 ]. About 1-2 mm of the sample area is then removed using a sterilized micropunch. Before it could be processed for further down-stream applications, the punch is washed with detergent and rinsed [ 14 , 43 ].
DNA extraction using cellulose-based paper is fast, highly convenient, does not require profound laboratory expertise and enables easy storage of the sample without refrigeration. However, downstream processing to recover pure and concentrated DNA can be very challenging [ 44 ].
Chelex-100 Extraction
In 2011, Xiong Hui, Xie Liqun and Chen Jiayi patented a DNA extraction method using chelex-100 [ 45 ]. Chelex resin is a styrenedivinylbenzene copolymer used to chelate metal ions that act as cofactors for DNases with its iminodiacetic acid groups. After overnight incubation, 5% chelex solution and proteinase K used to degrade the DNases are added. The tissue sample is then boiled in the 5% chelex solution to lyse the remaining membranes as well as to denature the proteins and DNA. Chelex prevents DNA from being digested by DNases that might remain after boiling, hence stabilizing the preparation. The resulting single-stranded DNA can then be concentrated from the supernatant after centrifugation [ 45 ].
Advantages of this method include the reduced risk for contamination and mis-pipetting since the procedure only involves the use of a single test tube and few manipulating steps [ 55 ]. However, the lack of purification renders this method ineffective in the removal of PCR inhibitors. Another limitation is that the isolated DNA can be unstable and is not suitable for RFLP analysis [ 46 ].
Filter paper-based DNA extraction method
In 2017, Rui Shi and Dilip Panthee describe a DNA extraction method using filter paper, which can be used to isolate DNA from plant sources. A spin plate composed of a 96-well plate (V-bottom) with a hole (about 1 mm in diameter) drilled into the bottom of each well is used, with each well containing a disc of Whatman™ filter paper (approximately 8 mm in diameter). Samples treated with lysis buffers are filtered with centrifugation. This method replaces glass fiber filters used in commercial kits for DNA extraction with filter paper, hence greatly reducing the costs of this method [ 47 ].
Since the first DNA extraction performed by Friedrich Miescher in 1869, scientists have made extraordinary progress in designing extraction methods that are more reliable, easier and faster to perform, more cost-effective and produce a higher yield. Newer techniques that are more reliable and efficient have facilitated the advancement in knowledge about the human genome and played a major role in the advent of various fields in science such as gene-editing and personalized medicine. However, at present it seems that there is no single procedure that is applicable to all contexts of DNA extraction due to their restrictions in producing yields with optimal purity and convenience of use. Hence, solutions to overcome the limitations of these techniques are required for better results and to simplify the handling of DNA. Improvement in the design of existing methods and developing new techniques will be the driving force to direct further development of future DNA extraction technology.
College of Medicine, Gulf Medical University, Ajman, UAE.
- Dahm R (2005) Friedrich Miescher and the discovery of DNA. Developmental Biology 278(2): 274-288 .
- Dhaliwal A (2013) DNA Extraction and Purification. Materials and Methods p. 3 .
- Bruce Alberts, Alexander Johnson, Julian Lewis, Martin Raff, Keith Roberts, et al. (2002) Molecular biology of the cell (4 th edn). Garland Science, New York, USA .
- Hardison R (2020) 2.5: B-Form, A-Form, and Z-Form of DNA.
- Eun H (1996) Enzymes and Nucleic Acids. Enzymology Primer for Recombinant DNA Technology, pp. 1-108.
- Modrow S, Falke D, Truyen U, Schatzl H Viruses with a Single-Stranded DNA Genome. Molecular Virology (1 st edition) Springer, Berlin, Germany , pp. 875-918.
- Popov L (1992) Single-stranded DNA from viruses, prokaryotes and eukaryotes. Molecular Biology 26(1): 25-58 .
- Reference G (2020) What is pharmacogenomics?
- Sirkov I (2016) Nucleic Acid Isolation and Downstream Applications. Nucleic Acids - From Basic Aspects to Laboratory Tools p. 1-26 .
- Evans J (2001) The complexities of predictive genetic testing. BMJ 322(7293): 1052-1056.
- Gaj T, Sirk S, Shui S, Liu J (2016) Genome-Editing Technologies: Principles and Applications. Cold Spring Harbor Perspectives in Biology 8(12) .
- Ali N, Rampazzo R, Costa A, Krieger M (2017) Current Nucleic Acid Extraction Methods and Their Implications to Point-of-Care Diagnostics. Biomed Research International.
- Griffiths L, Chacon Cortes D (2014) Methods for extracting genomic DNA from whole blood samples: current perspectives. Journal of Biorepository Science for Applied Medicine 2: 1-9 .
- Carpi FM, Di Pietro F, Vincenzetti S, Mignini F, Napolioni V (2011) Human DNA Extraction Methods: Patents and Applications. Recent Patents on DNA & Gene Sequences 5(1): 1-7 .
- (1955) Proceedings of the Biochemical Society. The Biochemical journal 60(4): xxxi–xlii.
- Peterson E, Sober H (1956) Chromatography of Proteins. I. Cellulose Ion-exchange Adsorbents. Journal of The American Chemical Society 78(4): 751-755 .
- Cuatrecasas P, Wilchek M, Anfinsen C (1968) Selective enzyme purification by affinity chromatography. Proceedings of The National Academy of Sciences 61(2): 636-643 .
- Tan S, Yiap B (2009) DNA, RNA, and Protein Extraction: The Past and The Present. Journal of Biomedicine and Biotechnology: 1-10
- Almeida A, Eusébio D, Queiroz J, Sousa F, Sousa A (2020) The use of size-exclusion chromatography in the isolation of supercoiled minicircle DNA from Escherichia coli lysate. Journal of Chromatography A 1609 .
- Budelier K, Schorr J (1998) Purification of DNA by Anion‐Exchange Chromatography. Current Protocols in Molecular Biology 42(1).
- Chockalingam P, Jurado L, Jarrett H (2001) DNA Affinity Chromatography. Molecular Biotechnology 19(2): 189-200.
- Meselson M, Stahl F, Vinograd J (1957) EQUILIBRIUM SEDIMENTATION OF MACROMOLECULES IN DENSITY GRADIENTS. Proceedings of The National Academy of Sciences 43(7): 581-588 .
- Leland J Cseke, Ara Kirakosyan, Peter B Kaufman, Margaret V Westfall (2012) Handbook of molecular and cellular methods in biology and medicine (3 rd Edition). Boca Raton: CRC Press/Taylor & Francis Group, UK .
- Birnboim H, Doly J (1979) A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Research 7(6): 1513-1523 .
- Vogelstein B, Gillespie D (1979) Preparative and analytical purification of DNA from agarose. Proceedings of The National Academy of Sciences 76(2): 615-619 .
- Höss M, Pääbo S (1993) DNA extraction from Pleistocene bones by a silica-based purification method. Nucleic Acids Research 21(16): 3913-3914 .
- Woodard D, Howard A, Down J (1993) PROCESS FOR PURIFYING DNA ON HYDRATED SILCA. United States.
- Esser K, Marx W, Lisowsky T (2006) maxXbond: first regeneration system for DNA binding silica matrices. Nature Methods 3(1) i-ii .
- Miller S, Dykes D, Polesky H (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Research 16(3): 1215.
- Javadi A, Shamaei M, Mohammadi Ziazi L, Pourabdollah M, Dorudinia A, et al. (2014) Qualification study of two genomic DNA extraction methods in different clinical samples. Tanaffos 13(4): 41-47.
- Doyle J (1991) DNA Protocols for Plants. Molecular Techniques in Taxonomy pp. 283-293 .
- Green M, Sambrook J (2020) Isolation of High-Molecular-Weight DNA Using Organic Solvents. CSH.
- Elkins K (2013) DNA Extraction. Forensic DNA Biology p. 39-52.
- Ebeling w, Hennrich N, Klockow M, Metz H, Orth H, et al. (1974) Proteinase K from Tritirachium album Limber. European Journal of Biochemistry 47(1): 91-97.
- Thomas S, Moreno R, Tilzer L (1989) DNA extraction with organic solvents in gel barrier tubes. Nucleic Acids Research 17(13): 5411-5411.
- Hawkins T (1998) DNA purification and isolation using magnetic particles. United States.
- Elkins K (2013) Forensic DNA biology, Oxford Academic Press, Kidlington, England, pp. 43-45.
- Saiyed Z, Ramchand C (2007) Extraction of Genomic DNA Using Magnetic Nanoparticles (Fe3O4) as a Solid-Phase Support. American Journal of Infectious Diseases 3(4): 225-229 .
- Abd El Aal A, Abd Elghany N, Mohamadin A, El Badry A (2010) Comparative study of five methods for DNA extraction from whole blood samples. IJHS, 3(1).
- Fomovskaia G, Smith M, Davis J, Jones K (2000) Fta-coated media for use as a molecular diagnostic tool, USA.
- (2007) Whatman™ FTA™ GE Healthcare Life Sciences.
- (2017) Whatman® FTA® card technology.
- Siegel C, Stevenson F, Zimmer E (2017) Evaluation and Comparison of FTA Card and CTAB DNA Extraction Methods for Non-Agricultural Taxa. Applications in Plant Sciences: 5(2).
- Hui X, Liqun X, Jiayi C (2014) Method for rapidly extracting nucleic acid from biological sample. China.
- Hoy M (2018) Insect molecular genetics. pp. 320.
- Shi R, Panthee D (2017) A novel plant DNA extraction method using filter paper-based 96-well spin plate. Planta 246(3): 579-584.

Sign up for Newsletter
Sign up for our newsletter to receive the latest updates. We respect your privacy and will never share your email address with anyone else.
American Journal of Biomedical Science & Research (ISSN: 2642-1747) is an Open access online Journal dedicated in advancing the latest scientific knowledge of science, medicine, technology and its related disciplines.
Contact info
BiomedGrid LLC, 333 City Boulevard West, 17 th Floor, Orange, California, 92868, USA +1 (626) 698-0574 [email protected]
Quick Links
Aim & scope
Editorial Committee
Open Access
Peer Review Process
Special Issues
Online Submission
Register for Editorial Committee
Recommended a Librarian
Advertise with us
Refer a friend.

Track Your Article
Suggested by
Referrer Details
Disseminate your Research today!
We offers a wealth of free resources on academic research and publishing. Sign up and get complete access to a vibrant global community of researchers.
If you forgot / lost your password, please click on this link to reset it.
Not a member? Sign Up
Forgot Password?
Already have an account? Log In
Unsubscribed
Your request for unsubscribing was successful. Unsubscription will be finalized in next 24 hours. During this time you may receive some previously planned communications.
Join as Reviewer We welcome academicians to join our committee

Home — Essay Samples — Science — DNA — Review of the Process of DNA Extraction
Review of The Process of DNA Extraction
- Categories: DNA Genetic Engineering Genetic Modification
About this sample

Words: 952 |
Published: Mar 3, 2020
Words: 952 | Pages: 2 | 5 min read
Table of contents
Introduction, usage of dna extraction.

Cite this Essay
To export a reference to this article please select a referencing style below:
Let us write you an essay from scratch
- 450+ experts on 30 subjects ready to help
- Custom essay delivered in as few as 3 hours
Get high-quality help

Dr. Karlyna PhD
Verified writer
- Expert in: Science
+ 120 experts online
By clicking “Check Writers’ Offers”, you agree to our terms of service and privacy policy . We’ll occasionally send you promo and account related email
No need to pay just yet!
Related Essays
1 pages / 517 words
2 pages / 804 words
1 pages / 674 words
2 pages / 914 words
Remember! This is just a sample.
You can get your custom paper by one of our expert writers.
121 writers online
Still can’t find what you need?
Browse our vast selection of original essay samples, each expertly formatted and styled
Related Essays on DNA
It is joining together of DNA molecules from two different species that are inserted into a host organism to produce new genetic combinations that are of value to science, medicine, agriculture, and industry. Since the focus of [...]
Polymerase chain reaction or PCR is the in vitro technique for the synthesis of the DNA or we can say that it is an in vitro DNA replication procedure. This technique is generally used in case of molecular biology for the [...]
Heredity is the transmission of genetic characteristics from ancestor to descendant through the genes. As a subject, it is tied closely to genetics, the area of biological study concerned with hereditary traits. The study of [...]
Beyond The Central Dogma The principal dogma of molecular biology explains that DNA codes for RNA, which codes for proteins. Within the crucial Dogma, you can find out about the vital roles of messenger RNA, switch RNA and [...]
Matthew Meselson and Franklin Stahl are famous for their DNA replication experiment. Meselson and Stahl conducted the experiment which supported the hypothesis made by Watson and Crick that DNA replicates. The purpose of the [...]
The 'scientific' theory of eugenics emerged in 1869 through Francis Galton. Galton, a hereditary genius and cousin to Charles Darwin, believed that behavior and characteristics were passed through generations hereditarily. For [...]
Related Topics
By clicking “Send”, you agree to our Terms of service and Privacy statement . We will occasionally send you account related emails.
Where do you want us to send this sample?
By clicking “Continue”, you agree to our terms of service and privacy policy.
Be careful. This essay is not unique
This essay was donated by a student and is likely to have been used and submitted before
Download this Sample
Free samples may contain mistakes and not unique parts
Sorry, we could not paraphrase this essay. Our professional writers can rewrite it and get you a unique paper.
Please check your inbox.
We can write you a custom essay that will follow your exact instructions and meet the deadlines. Let's fix your grades together!
Get Your Personalized Essay in 3 Hours or Less!
We use cookies to personalyze your web-site experience. By continuing we’ll assume you board with our cookie policy .
- Instructions Followed To The Letter
- Deadlines Met At Every Stage
- Unique And Plagiarism Free
The current status and trends of DNA extraction
Affiliations.
- 1 Department of Chemistry, Faculty of Science, Hong Kong Baptist University (HKBU), Hong Kong, Hong Kong.
- 2 Division of Science and Technology, Beijing Normal University - Hong Kong Baptist University United International College (UIC), Zhuhai, Guangdong, China.
- PMID: 37338306
- DOI: 10.1002/bies.202200242
DNA extraction, playing an irreplaceable role in molecular biology as it is an essential step prior to various downstream biological analyses. Thus, the accuracy and reliability of downstream research outcomes depend largely on upstream DNA extraction methodology. However, with the advancement of downstream DNA detection techniques, the development of corresponding DNA extraction methods is lagging behind. The most innovative DNA extraction techniques are silica- or magnetic-based. Recent studies have demonstrated that plant fiber-based adsorbents (PF-BAs) have stronger DNA capturing ability than classic materials. Moreover, magnetic ionic liquid (MIL)-based DNA extraction has gathered attention lately, and extrachromosomal circular DNA (eccDNA), cell-free DNA (cfDNA), and microbial community DNA are current research hotspots. These require specific extraction methods, along with constant improvements in the way they are used. This review discusses the significance as well as the direction of innovation of DNA extraction methods to try to provide valuable references including current status and trends for DNA extraction.
Keywords: cell-free DNA; extrachromosomal circular DNA; magnetic ionic liquid; microbial community; plant fiber-based adsorbent.
© 2023 Wiley Periodicals LLC.
Publication types
- Ionic Liquids*
- Reproducibility of Results
- Ionic Liquids

IMAGES
VIDEO
COMMENTS
Keywords: DNA extraction, Polymerase chain reaction, real time PCR. DNA E XTRACTION. DNA extraction is a method to purify DNA by using physical and/or chemical methods from a sample separating DNA from cell membranes, proteins, and other cellular components. Friedrich Miescher in 1869 did DNA isolation for the first time.
Enzymatic method of DNA extraction: This method is actually a combination of a salt method and an enzymatic method. Before proceeding with the enzymatic digestion, the extraction buffer is used. The extraction buffer's composition varies each lab, but the main ingredients are Tris, EDTA, NaCl, sodium lauryl sulfate, and SDS. No phenol ...
Extraction and analytical methods for the identification of parasites in food. In Advanced Food Analysis Tools, 2021. DNA extraction. DNA extraction is the process where DNA is separated from proteins, membranes, and other cellular material (Butler, 2012).According to Rice (2018), the method involves three necessary steps, namely, lysed, precipitation, and purification.
The DNA extraction process involves disrupting cell membranes to release genetic material, isolating and purifying DNA from other cellular components, and then concentrating and purifying the DNA itself. ... Whatman No. 1 filter papers were immersed in an infected leaf extract before washing once, twice, or not at all before amplification ...
DNA extraction Definition. DNA extraction is the technique used to isolate DNA in a biological sample. As shown in this photo, DNA, a long stringy molecule, can be lifted out of a solution by the use of a glass rod or wooden stick which it naturally wraps around when turned. Importance. The extraction of DNA is pivotal to biotechnology.
DNA Extraction Methods. John M. Butler, in Advanced Topics in Forensic DNA Typing: Methodology, 2012 Protocols for Various Tissue Types. Optimal DNA extraction may come from slightly different procedures depending on the source of the biological sample. The reference lists at the back of this chapter include citations to methods developed for DNA extraction from a variety of tissue types ...
Since the first DNA extraction performed by Friedrich Miescher in 1869, scientists have made extraordinary progress in designing extraction methods that are more reliable, easier and faster to perform, more cost-effective and produce a higher yield. The classic liquid-liquid DNA extraction method involves the use of organic and inorganic reagents such as phenol-chloroform which pose a toxic ...
DNA extraction is the process in which DNA is separated from the cell material contained in the cell acquired. Although this process is immensely useful in genetics research and forensic testing, it is a very critical and labor-intensive process.
DNA extraction, playing an irreplaceable role in molecular biology as it is an essential step prior to various downstream biological analyses. Thus, the accuracy and reliability of downstream research outcomes depend largely on upstream DNA extraction methodology. However, with the advancement of do …
Abstract DNA extraction, playing an irreplaceable role in molecular biology as it is an essential step prior to various downstream biological analyses. ... Search for more papers by this author. Bo Lei, Corresponding Author. Bo Lei [email protected] Division of Science and Technology, Beijing Normal University - Hong Kong Baptist University ...